[English] 日本語
 Yorodumi
Yorodumi- EMDB-25487: SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (l... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Spike NTD in complex with neutralizing Fab SARS2-57 (local refinement) | |||||||||
 Map data Map data | Local reconstruction of SARS-CoV-2 Spike NTD in complex with Fab SARS2-57 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycoprotein / Antibody / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID / VIRAL PROTEIN-IMMUNE SYSTEM complex / Center for Structural Biology of Infectious Diseases / CSBID | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
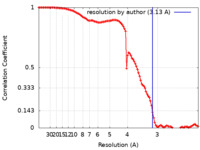
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Adams LJ / Fremont DH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2023 Journal: Cell Rep Med / Year: 2023Title: A broadly reactive antibody targeting the N-terminal domain of SARS-CoV-2 spike confers Fc-mediated protection. Authors: Lucas J Adams / Laura A VanBlargan / Zhuoming Liu / Pavlo Gilchuk / Haiyan Zhao / Rita E Chen / Saravanan Raju / Zhenlu Chong / Bradley M Whitener / Swathi Shrihari / Prashant N Jethva / ...Authors: Lucas J Adams / Laura A VanBlargan / Zhuoming Liu / Pavlo Gilchuk / Haiyan Zhao / Rita E Chen / Saravanan Raju / Zhenlu Chong / Bradley M Whitener / Swathi Shrihari / Prashant N Jethva / Michael L Gross / James E Crowe / Sean P J Whelan / Michael S Diamond / Daved H Fremont /  Abstract: Most neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) target the receptor binding domain (RBD) of the spike (S) protein. Here, we characterize a panel of mAbs targeting the N-terminal ...Most neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) target the receptor binding domain (RBD) of the spike (S) protein. Here, we characterize a panel of mAbs targeting the N-terminal domain (NTD) or other non-RBD epitopes of S. A subset of NTD mAbs inhibits SARS-CoV-2 entry at a post-attachment step and avidly binds the surface of infected cells. One neutralizing NTD mAb, SARS2-57, protects K18-hACE2 mice against SARS-CoV-2 infection in an Fc-dependent manner. Structural analysis demonstrates that SARS2-57 engages an antigenic supersite that is remodeled by deletions common to emerging variants. In neutralization escape studies with SARS2-57, this NTD site accumulates mutations, including a similar deletion, but the addition of an anti-RBD mAb prevents such escape. Thus, our study highlights a common strategy of immune evasion by SARS-CoV-2 variants and how targeting spatially distinct epitopes, including those in the NTD, may limit such escape. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25487.map.gz emd_25487.map.gz | 90.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25487-v30.xml emd-25487-v30.xml emd-25487.xml emd-25487.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25487_fsc.xml emd_25487_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25487.png emd_25487.png | 70.1 KB | ||
| Filedesc metadata |  emd-25487.cif.gz emd-25487.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25487 http://ftp.pdbj.org/pub/emdb/structures/EMD-25487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25487 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25487 | HTTPS FTP |
-Related structure data
| Related structure data |  7swwMC  7swxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25487.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25487.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local reconstruction of SARS-CoV-2 Spike NTD in complex with Fab SARS2-57 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 spike bound by SARS2-57 Fab
| Entire | Name: SARS-CoV-2 spike bound by SARS2-57 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike bound by SARS2-57 Fab
| Supramolecule | Name: SARS-CoV-2 spike bound by SARS2-57 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.082352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLK UniProtKB: Spike glycoprotein |
-Macromolecule #2: SARS2-57 Fv heavy chain
| Macromolecule | Name: SARS2-57 Fv heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.955321 KDa |
| Sequence | String: EVQLQQSGAE LVRPGALVKL SCKASGFNIK DYFVHWVKQR PVQGLEWIGW IDPENGNTIY GPKFQGKASL AADTSSNTGY LQLSSLTSE DTAVYYCARW DGYETLDYWG QGTSVTVS |
-Macromolecule #3: SARS2-57 Fv light chain
| Macromolecule | Name: SARS2-57 Fv light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.383759 KDa |
| Sequence | String: DIVMSQSPSS LAVSVGEKVT MSCQSSQSLL YSNNEKNYLA WYQQKPGHSP KLLLYWASTR ESGVPDRFTG SGSGTAFTLT ISSVKAEDL AVYYCQQYYN YPYTFGGGTK LEI |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 66.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)