[English] 日本語
 Yorodumi
Yorodumi- EMDB-25488: SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Spike in complex with neutralizing Fab SARS2-57 (three down conformation) | |||||||||
 Map data Map data | Density map of SARS-CoV-2 spike bound by SARS2-57 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycoprotein / Antibody / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID / VIRAL PROTEIN-IMMUNE SYSTEM complex / Center for Structural Biology of Infectious Diseases / CSBID | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
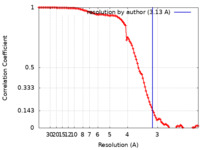
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Adams LJ / Fremont DH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2023 Journal: Cell Rep Med / Year: 2023Title: A broadly reactive antibody targeting the N-terminal domain of SARS-CoV-2 spike confers Fc-mediated protection. Authors: Lucas J Adams / Laura A VanBlargan / Zhuoming Liu / Pavlo Gilchuk / Haiyan Zhao / Rita E Chen / Saravanan Raju / Zhenlu Chong / Bradley M Whitener / Swathi Shrihari / Prashant N Jethva / ...Authors: Lucas J Adams / Laura A VanBlargan / Zhuoming Liu / Pavlo Gilchuk / Haiyan Zhao / Rita E Chen / Saravanan Raju / Zhenlu Chong / Bradley M Whitener / Swathi Shrihari / Prashant N Jethva / Michael L Gross / James E Crowe / Sean P J Whelan / Michael S Diamond / Daved H Fremont /  Abstract: Most neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) target the receptor binding domain (RBD) of the spike (S) protein. Here, we characterize a panel of mAbs targeting the N-terminal ...Most neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) target the receptor binding domain (RBD) of the spike (S) protein. Here, we characterize a panel of mAbs targeting the N-terminal domain (NTD) or other non-RBD epitopes of S. A subset of NTD mAbs inhibits SARS-CoV-2 entry at a post-attachment step and avidly binds the surface of infected cells. One neutralizing NTD mAb, SARS2-57, protects K18-hACE2 mice against SARS-CoV-2 infection in an Fc-dependent manner. Structural analysis demonstrates that SARS2-57 engages an antigenic supersite that is remodeled by deletions common to emerging variants. In neutralization escape studies with SARS2-57, this NTD site accumulates mutations, including a similar deletion, but the addition of an anti-RBD mAb prevents such escape. Thus, our study highlights a common strategy of immune evasion by SARS-CoV-2 variants and how targeting spatially distinct epitopes, including those in the NTD, may limit such escape. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25488.map.gz emd_25488.map.gz | 90.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25488-v30.xml emd-25488-v30.xml emd-25488.xml emd-25488.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25488_fsc.xml emd_25488_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25488.png emd_25488.png | 73 KB | ||
| Filedesc metadata |  emd-25488.cif.gz emd-25488.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25488 http://ftp.pdbj.org/pub/emdb/structures/EMD-25488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25488 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25488 | HTTPS FTP |
-Related structure data
| Related structure data |  7swxMC  7swwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25488.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25488.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of SARS-CoV-2 spike bound by SARS2-57 Fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 spike bound by SARS2-57 Fab
| Entire | Name: SARS-CoV-2 spike bound by SARS2-57 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike bound by SARS2-57 Fab
| Supramolecule | Name: SARS-CoV-2 spike bound by SARS2-57 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.519125 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NIT NLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPG QTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC YFPLQ SYGF QPTNGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQQFGR DIADTT DAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQDVN CTEVPVAIHA DQLTPTWRVY STGSNVFQTR AGCLIGA EH VNNSYECDIP IGAGICASYQ TQTNSPRRAR SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVTTE ILPVSMTK T SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFSQ ILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGPA LQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NQNAQALNTL VKQLSSNFGA I SSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FCGKGYHLMS FP QSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIV NNTVYD PLQPELD UniProtKB: Spike glycoprotein |
-Macromolecule #2: SARS2-57 Fv heavy chain
| Macromolecule | Name: SARS2-57 Fv heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.955321 KDa |
| Sequence | String: EVQLQQSGAE LVRPGALVKL SCKASGFNIK DYFVHWVKQR PVQGLEWIGW IDPENGNTIY GPKFQGKASL AADTSSNTGY LQLSSLTSE DTAVYYCARW DGYETLDYWG QGTSVTVS |
-Macromolecule #3: SARS2-57 Fv light chain
| Macromolecule | Name: SARS2-57 Fv light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.383759 KDa |
| Sequence | String: DIVMSQSPSS LAVSVGEKVT MSCQSSQSLL YSNNEKNYLA WYQQKPGHSP KLLLYWASTR ESGVPDRFTG SGSGTAFTLT ISSVKAEDL AVYYCQQYYN YPYTFGGGTK LEI |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 25 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 66.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)