[English] 日本語
 Yorodumi
Yorodumi- EMDB-27765: Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmet... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit) | |||||||||
 Map data Map data | Chikungunya VLP in complex with neutralizing Fab 506.C01 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody / VIRAL PROTEIN-IMMUNE SYSTEM complex / CSGID / Structural Genomics / Center for Structural Genomics of Infectious Diseases | |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane ...togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  Chikungunya virus strain Senegal 37997 / Chikungunya virus strain Senegal 37997 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Adams LJ / Fremont DH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2023 Journal: Sci Transl Med / Year: 2023Title: A chikungunya virus-like particle vaccine induces broadly neutralizing and protective antibodies against alphaviruses in humans. Authors: Saravanan Raju / Lucas J Adams / James T Earnest / Kelly Warfield / Lo Vang / James E Crowe / Daved H Fremont / Michael S Diamond /  Abstract: Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes epidemics of acute and chronic musculoskeletal disease. Here, we analyzed the human B cell response to a CHIKV-like particle- ...Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes epidemics of acute and chronic musculoskeletal disease. Here, we analyzed the human B cell response to a CHIKV-like particle-adjuvanted vaccine (PXVX0317) from samples obtained from a phase 2 clinical trial in humans (NCT03483961). Immunization with PXVX0317 induced high levels of neutralizing antibody in serum against CHIKV and circulating antigen-specific B cells up to 6 months after immunization. Monoclonal antibodies (mAbs) generated from peripheral blood B cells of three PXVX0317-vaccinated individuals on day 57 after immunization potently neutralized CHIKV infection, and a subset of these inhibited multiple related arthritogenic alphaviruses. Epitope mapping and cryo-electron microscopy defined two broadly neutralizing mAbs that uniquely bind to the apex of the B domain of the E2 glycoprotein. These results demonstrate the inhibitory breadth and activity of the human B cell response induced by the PXVX0317 vaccine against CHIKV and potentially other related alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27765.map.gz emd_27765.map.gz | 143.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27765-v30.xml emd-27765-v30.xml emd-27765.xml emd-27765.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27765_fsc.xml emd_27765_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27765.png emd_27765.png | 32.3 KB | ||
| Filedesc metadata |  emd-27765.cif.gz emd-27765.cif.gz | 7 KB | ||
| Others |  emd_27765_additional_1.map.gz emd_27765_additional_1.map.gz emd_27765_half_map_1.map.gz emd_27765_half_map_1.map.gz emd_27765_half_map_2.map.gz emd_27765_half_map_2.map.gz | 80.8 MB 151.8 MB 151.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27765 http://ftp.pdbj.org/pub/emdb/structures/EMD-27765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27765 | HTTPS FTP |
-Related structure data
| Related structure data |  8dwxMC  8dwwC  8dwyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27765.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27765.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chikungunya VLP in complex with neutralizing Fab 506.C01 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
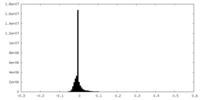
-Additional map: Unsharpened map
| File | emd_27765_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
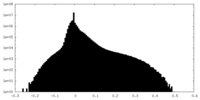
-Half map: Half map A
| File | emd_27765_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
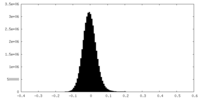
-Half map: Half map B
| File | emd_27765_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmet...
| Entire | Name: Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit) |
|---|---|
| Components |
|
-Supramolecule #1: Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmet...
| Supramolecule | Name: Chikungunya VLP in complex with neutralizing Fab 506.C01 (asymmetric unit) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 Chikungunya virus strain Senegal 37997 |
-Macromolecule #1: E1 glycoprotein
| Macromolecule | Name: E1 glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 47.346715 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHVTVIPNT VGVPYKTLVN RPGYSPMVLE MELQSVTLEP TLSLDYITCE YKTVIPSPYV KCCGTAECKD KSLPDYSCKV FTGVYPFMW GGAYCFCDAE NTQLSEAHVE KSESCKTEFA SAYRAHTASA SAKLRVLYQG NNITVAAYAN GDHAVTVKDA K FVVGPMSS ...String: YEHVTVIPNT VGVPYKTLVN RPGYSPMVLE MELQSVTLEP TLSLDYITCE YKTVIPSPYV KCCGTAECKD KSLPDYSCKV FTGVYPFMW GGAYCFCDAE NTQLSEAHVE KSESCKTEFA SAYRAHTASA SAKLRVLYQG NNITVAAYAN GDHAVTVKDA K FVVGPMSS AWTPFDNKIV VYKGDVYNMD YPPFGAGRPG QFGDIQSRTP ESKDVYANTQ LVLQRPAAGT VHVPYSQAPS GF KYWLKER GASLQHTAPF GCQIATNPVR AVNCAVGNIP ISIDIPDAAF TRVVDAPSVT DMSCEVPACT HSSDFGGVAI IKY TASKKG KCAVHSMTNA VTIREADVEV EGNSQLQISF STALASAEFR VQVCSTQVHC AAACHPPKDH IVNYPASHTT LGVQ DISTT AMSWVQKITG GVGLIVAVAA LILIVVLCVS FSRH UniProtKB: Structural polyprotein |
-Macromolecule #2: E2 glycoprotein
| Macromolecule | Name: E2 glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 46.858312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NFNVYKATRP YLAHCPDCGE GHSCHSPIAL ERIRNEATDG TLKIQVSLQI GIKTDDSHDW TKLRYMDSHT PADAERAGLL VRTSAPCTI TGTMGHFILA RCPKGETLTV GFTDSRKISH TCTHPFHHEP PVIGRERFHS RPQHGKELPC STYVQSTAAT A EEIEVHMP ...String: NFNVYKATRP YLAHCPDCGE GHSCHSPIAL ERIRNEATDG TLKIQVSLQI GIKTDDSHDW TKLRYMDSHT PADAERAGLL VRTSAPCTI TGTMGHFILA RCPKGETLTV GFTDSRKISH TCTHPFHHEP PVIGRERFHS RPQHGKELPC STYVQSTAAT A EEIEVHMP PDTPDRTLMT QQSGNVKITV NGQTVRYKCN CGGSNEGLTT TDKVINNCKI DQCHAAVTNH KNWQYNSPLV PR NAELGDR KGKIHIPFPL ANVTCRVPKA RNPTVTYGKN QVTMLLYPDH PTLLSYRNMG QEPNYHEEWV THKKEVTLTV PTE GLEVTW GNNEPYKYWP QMSTNGTAHG HPHEIILYYY ELYPTMTVVI VSVASFVLLS MVGTAVGMCV CARRRCITPY ELTP GATVP FLLSLLCCVR TTKA UniProtKB: Structural polyprotein |
-Macromolecule #3: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chikungunya virus strain Senegal 37997 / Strain: 37997 Chikungunya virus strain Senegal 37997 / Strain: 37997 |
| Molecular weight | Theoretical: 16.458701 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NDCIFEVKHE GKVMGYACLV GDKVMKPAHV KGTIDNADLA KLAFKRSSKY DLECAQIPVH MKSDASKFTH EKPEGYYNWH HGAVQYSGG RFTIPTGAGK PGDSGRPIFD NKGRVVAIVL GGANEGARTA LSVVTWNKDI VTKITPEGAE EW UniProtKB: Structural polyprotein |
-Macromolecule #4: 506.C01 light chain
| Macromolecule | Name: 506.C01 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.486756 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS VSASVGDRVT ITCRASQGIS SWLGWYQQKP GKAPKLLIYA ASSLQSGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ QANSFPRTFG QGTKVEIK |
-Macromolecule #5: 506.C01 heavy chain
| Macromolecule | Name: 506.C01 heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.0827 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSQTLSL TCTVSGGSIS SDDYYWTWIR LPPGKGLEWI GYIFYTGGTY YNPSLKSRVT ISLDRSKNQF SLKLSSVTA ADTAVYFCAR APETYCSTTN CYKGYFDSWG QGTLVTVSS |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 38.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 59000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)