-Search query
-Search result
Showing 1 - 50 of 1,074 items for (author: feng & ll)

EMDB-18878: 
Cryo-EM structure of the Asgard archaeal Argonaute HrAgo1 bound to a guide RNA

PDB-8r3z: 
Cryo-EM structure of the Asgard archaeal Argonaute HrAgo1 bound to a guide RNA

EMDB-42287: 
Cryo-EM map of human clmap-clamp loader ATAD5-RFC-gapped PCNA complex in intermediate state 3

EMDB-42288: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-two PCNAs complex in intermediate state 3

EMDB-42289: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-cracked PCNA complex in intermediate state 2

EMDB-42295: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-closed PCNA complex in intermediate state 1

PDB-8ui7: 
Cryo-EM map of human clmap-clamp loader ATAD5-RFC-gapped PCNA complex in intermediate state 3

PDB-8ui8: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-two PCNAs complex in intermediate state 3

PDB-8ui9: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-cracked PCNA complex in intermediate state 2

PDB-8uii: 
Cryo-EM map of human clamp-clamp loader ATAD5-RFC-closed PCNA complex in intermediate state 1

EMDB-41252: 
Cryo-EM map of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex

EMDB-41253: 
Cryo-EM map of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA

EMDB-41254: 
Cryo-EM map of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA

PDB-8thb: 
Structure of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex

PDB-8thc: 
Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA

PDB-8thd: 
Structure of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA
EMDB-17779: 
Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1

PDB-8pn9: 
Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1
EMDB-42383: 
Cryo-EM map of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2)
EMDB-42384: 
Cryo-EM map of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3)
EMDB-42385: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state I (state 5)
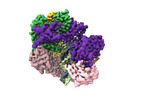
EMDB-42386: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex in the five-subunit binding state (state 4)
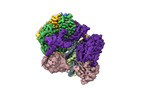
EMDB-42388: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state II (state 6)
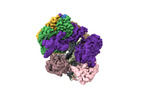
EMDB-42389: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with cracked and closed PCNA (state 7)

EMDB-42406: 
Cryo-EM map of the human CTF18-RFC alone in the apo state (State 1)

PDB-8umt: 
Atomic model of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2)

PDB-8umu: 
Atomic model of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3)

PDB-8umv: 
Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state I (state 5)

PDB-8umw: 
Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex in the five-subunit binding state (state 4)

PDB-8umy: 
Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state II (state 6)

PDB-8un0: 
Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with cracked and closed PCNA (state 7)

PDB-8unj: 
Atomic model of the human CTF18-RFC alone in the apo state (State 1)

EMDB-38284: 
XBB.1.5 spike protein in complex with BD55-1205

EMDB-36858: 
Structure of PKD2-F604P complex

PDB-8k3s: 
Structure of PKD2-F604P complex

EMDB-34848: 
Structure of PKD2-F604P (Polycystin-2, TRPP2) with ML-SA1

PDB-8hk7: 
Structure of PKD2-F604P (Polycystin-2, TRPP2) with ML-SA1

EMDB-18267: 
Structure of the human 20S U5 snRNP core

EMDB-19041: 
Structure of the human 20S U5 snRNP

PDB-8q91: 
Structure of the human 20S U5 snRNP core

PDB-8rc0: 
Structure of the human 20S U5 snRNP

EMDB-17509: 
Cryo-EM structure of CAK in complex with inhibitor BS-181

EMDB-17510: 
Cryo-EM structure of CAK in complex with inhibitor BS-194

EMDB-17512: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0510-R

EMDB-17513: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0510-S

EMDB-17514: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0574

EMDB-17515: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0768

EMDB-17516: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0829

EMDB-17517: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0880 (ring-up conformation)

EMDB-17518: 
Cryo-EM structure of CAK in complex with inhibitor ICEC0880 (ring-down conformation)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
