-Search query
-Search result
Showing all 42 items for (author: dietrich & ka)

EMDB-70812: 
Tetrameric POLQ Helicase-like Domain Bound to Cmpd 19, a Small-Molecule ATPase Inhibitor and Drug Candidate Analog
Method: single particle / : Zahn KE, Scapin G

EMDB-70813: 
Tetrameric POLQ Helicase-like Domain Bound to Cmpd 36, a Small-Molecule ATPase Inhibitor and Drug Candidate Analog
Method: single particle / : Zahn KE, Scapin G

PDB-9osw: 
Tetrameric POLQ Helicase-like Domain Bound to Cmpd 19, a Small-Molecule ATPase Inhibitor and Drug Candidate Analog
Method: single particle / : Zahn KE, Mader P, Sicheri F

PDB-9osy: 
Tetrameric POLQ Helicase-like Domain Bound to Cmpd 36, a Small-Molecule ATPase Inhibitor and Drug Candidate Analog
Method: single particle / : Zahn KE, Mader P, Sicheri F

EMDB-48672: 
Consensus map of the endogenous Pfs230-Pfs48/45 complex
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

EMDB-48669: 
Focused map of Pfs230 domains 1-8 of the endogenous Pfs230-Pfs48/45 complex
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

EMDB-48670: 
Focused map of Pfs230 (domains 9-14) and Pfs48/45 of the endogenous Pfs230-Pfs48/45 complex
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

EMDB-48673: 
Composite map of the endogenous complex of Pfs230-Pfs48/45
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

PDB-9mvt: 
Pfs230 domains 1-8 of the endogenous Pfs230-Pfs48/45 complex
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

PDB-9mvv: 
Pfs230 (D9-D14) with Pfs48/45 of the endogenous Pfs230-Pfs48/45 complex
Method: single particle / : Dietrich MH, Glukhova A, Shakeel S, Tham WH

EMDB-13246: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila
Method: electron tomography / : Boeck D, Huesler D

EMDB-13247: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila
Method: electron tomography / : Boeck D, Huesler D

EMDB-13248: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila
Method: electron tomography / : Boeck D, Huesler D

EMDB-13249: 
cryoFIB milling/cryoET of amoeba infected with Legionella pneumophila
Method: electron tomography / : Boeck D, Huesler D

EMDB-24642: 
SARS-CoV-2 Spike bound to Fab PDI 210
Method: single particle / : Pymm P, Glukhova A, Black K, Tham WH

EMDB-24643: 
SARS-CoV-2 Spike bound to Fab PDI 96
Method: single particle / : Pymm P, Glukhova A, Black K, Tham WH

EMDB-24644: 
SARS-CoV-2 Spike bound to Fab PDI 215
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24645: 
SARS-CoV-2 Spike bound to Fab WCSL 119
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24646: 
SARS-CoV-2 Spike bound to Fab WCSL 129
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24647: 
SARS-CoV-2 Spike bound to Fab PDI 93
Method: single particle / : Black K, Glukhova A, Pymm P, Tham WH

EMDB-24648: 
SARS-CoV-2 Spike bound to Fab PDI 222
Method: single particle / : Glukhova A, Pymm P, Black K, Tham WH

EMDB-24649: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222
Method: single particle / : Pymm P, Glukhova A

PDB-7rr0: 
SARS-CoV-2 receptor binding domain bound to Fab PDI 222
Method: single particle / : Pymm P, Glukhova A, Black KA, Tham WH

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase
Method: subtomogram averaging / : Dietrich L

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria
Method: helical / : Parey K

EMDB-23566: 
The SARS-CoV-2 spike protein receptor binding domain bound to neutralizing nanobodies WNb 2 and WNb 10
Method: single particle / : Pymm P, Glukhova A

EMDB-23567: 
Cryo-EM map of SARS-CoV-2 Spike protein bound to neutralising nanobodies WNb2 and WNb10 (C3 symmetry)
Method: single particle / : Pymm P, Tham WH, Glukhova A

EMDB-23568: 
Cryo-EM map of SARS-CoV-2 Spike protein in complex with neutralising nanobodies WNb2 and WNb10 (C1 symmetry)
Method: single particle / : Pymm P, Tham WH, Glukhova A

PDB-7lx5: 
The SARS-CoV-2 spike protein receptor binding domain bound to neutralizing nanobodies WNb 2 and WNb 10
Method: single particle / : Pymm P, Glukhova A, Tham WH

EMDB-11997: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

PDB-7b3y: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
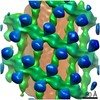
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M
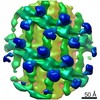
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O
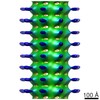
EMDB-2094: 
Central hub structure of Trichonympha cartwheel
Method: subtomogram averaging / : Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P

EMDB-5011: 
Crosslinked kinesin head-tail complex bound to the microtubule
Method: helical / : Dietrich KA, Sindelar CV, Brewer PD, Downing KH, Cremo CR, Rice SE
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
