-Search query
-Search result
Showing 1 - 50 of 613 items for (author: burg & m)

EMDB-43296: 
Constituent EM map: Focused refinement S100A1 of mouse RyR1 in complex with S100A1 (EGTA-only dataset)
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry
EMDB-43712: 
Human EBP complexed with compound 1
EMDB-43713: 
Human EBP complexed with compound 3a

PDB-8w0r: 
Human EBP complexed with compound 1

PDB-8w0s: 
Human EBP complexed with compound 3a

EMDB-17377: 
Structure of human SIT1 (focussed map / refinement)

EMDB-17378: 
Structure of human SIT1:ACE2 complex (open PD conformation)

EMDB-17379: 
Structure of human SIT1:ACE2 complex (closed PD conformation)

EMDB-17380: 
Structure of human SIT1 bound to L-pipecolate (focussed map / refinement)

EMDB-17381: 
Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate

EMDB-17382: 
Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate

PDB-8p2w: 
Structure of human SIT1 (focussed map / refinement)

PDB-8p2x: 
Structure of human SIT1:ACE2 complex (open PD conformation)

PDB-8p2y: 
Structure of human SIT1:ACE2 complex (closed PD conformation)

PDB-8p2z: 
Structure of human SIT1 bound to L-pipecolate (focussed map / refinement)

PDB-8p30: 
Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate

PDB-8p31: 
Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate

EMDB-50356: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-50357: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry
EMDB-50359: 
Rhodobacter microvirus Ebor attached to B10 host cell reconstructed by single particle analysis with applied C5 symmetry
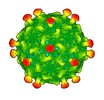
EMDB-50360: 
Rhodobacter microvirus Ebor attached to the outer membrane vesicle

EMDB-50361: 
Rhodobacter microvirus Ebor attached to the host cell reconstructed by subtomogram averaging

PDB-9ffg: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

PDB-9ffh: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-17197: 
Human TPC2 in Complex with Antagonist (S)-SG-094

EMDB-19108: 
Human TPC2 in Complex withAntagonist (R)-SG-094

PDB-8ouo: 
Human TPC2 in Complex with Antagonist (S)-SG-094
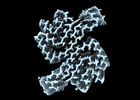
EMDB-19986: 
Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo

PDB-9euu: 
Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo

EMDB-41271: 
Consensus map of 96nm repeat of human respiratory doublet microtubule, RS1-2 region

EMDB-19408: 
Cryo-EM structure of CDK2-cyclin A in complex with CDC25A

EMDB-41152: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation

PDB-8tc5: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation

EMDB-18729: 
Cryo-EM structure of tetrameric human SAMHD1 with dApNHpp

EMDB-18730: 
Cryo-EM structure of tetrameric human SAMHD1 State I - Tense

EMDB-18731: 
Cryo-EM structure of tetrameric human SAMHD1 State II - Hemi-relaxed

EMDB-18732: 
Cryo-EM structure of tetrameric human SAMHD1 State III - Relaxed

EMDB-18733: 
Cryo-EM structure of tetrameric human SAMHD1 State IV - Depleted relaxed

EMDB-18734: 
Cryo-EM structure of tetrameric human SAMHD1 State V - Depleted relaxed

PDB-8qxj: 
Cryo-EM structure of tetrameric human SAMHD1 with dApNHpp

PDB-8qxk: 
Cryo-EM structure of tetrameric human SAMHD1 State I - Tense

PDB-8qxl: 
Cryo-EM structure of tetrameric human SAMHD1 State II - Hemi-relaxed

PDB-8qxm: 
Cryo-EM structure of tetrameric human SAMHD1 State III - Relaxed

PDB-8qxn: 
Cryo-EM structure of tetrameric human SAMHD1 State IV - Depleted relaxed

PDB-8qxo: 
Cryo-EM structure of tetrameric human SAMHD1 State V - Depleted relaxed

EMDB-41149: 
Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation

EMDB-41150: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation

PDB-8tc0: 
Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation

PDB-8tc1: 
Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
