-Search query
-Search result
Showing 1 - 50 of 376 items for (author: brandt & j)

EMDB-16158: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 peripheral arm)

EMDB-16159: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 CIII)

EMDB-16160: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 membrane arm)

EMDB-16161: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 peripheral arm)

EMDB-16163: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 CIII)

EMDB-42990: 
DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase

EMDB-42991: 
DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase

EMDB-42992: 
DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase

EMDB-42993: 
DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v5m: 
Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v5n: 
Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v5o: 
Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v6g: 
DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v6h: 
DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v6i: 
DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase

PDB-8v6j: 
DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase

EMDB-42034: 
RNA priming complex of Human polymerase alpha-primase (Conformation 2)

EMDB-42036: 
Human Polymerase alpha-Primase, polymerase alpha catalytic domain deletion mutant (Conformation 1)
EMDB-42037: 
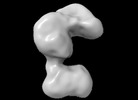
Human Polymerase alpha-Primase, polymerase alpha catalytic domain deletion mutant (Conformation 2)
EMDB-42033: 
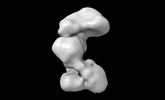
RNA priming complex of Human Polymerase-Alpha-Primase (Conformation 1)
EMDB-42035: 
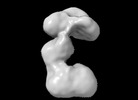
RNA priming complex of Human Polymerase alpha-primase (Conformation 3)

EMDB-42140: 
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase

EMDB-42141: 
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase

EMDB-42142: 
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase

PDB-8ucu: 
Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase

PDB-8ucv: 
Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase

PDB-8ucw: 
Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase

EMDB-15849: 
Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom

EMDB-15850: 
Cryo-EM structure of the Neurospora crassa TOM core complex with Tom20 over one pore

EMDB-15856: 
Cryo-EM structure of the Neurospora crassa TOM core complex with Tom20 over both pores

PDB-8b4i: 
Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom
EMDB-15335: 
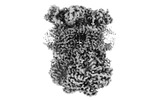
Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, Rieske domains in b- and c-position
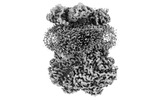
EMDB-15338: 
Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, Rieske domains in the int-position
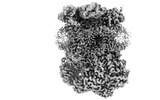
EMDB-15337: 
Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, Rieske domains in the c-position
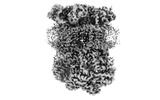
EMDB-15336: 
Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, Rieske domains in b-position

EMDB-16172: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition)

PDB-8bq6: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition)

EMDB-29862: 
Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase

EMDB-29864: 
Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase

EMDB-29871: 
DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase

EMDB-29872: 
Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase

EMDB-29873: 
Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase

EMDB-29888: 
Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase

EMDB-29889: 
Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase

EMDB-29891: 
Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase

PDB-8g99: 
Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase

PDB-8g9f: 
Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase

PDB-8g9l: 
DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase

PDB-8g9n: 
Partial DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase

PDB-8g9o: 
Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
