[English] 日本語
 Yorodumi
Yorodumi- EMDB-42035: RNA priming complex of Human Polymerase alpha-primase (Conformation 3) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA priming complex of Human Polymerase alpha-primase (Conformation 3) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Polymerase / Primase / Polymerase Alpha / RNA priming / REPLICATION | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA primase AEP / ribonucleotide binding / DNA replication initiation / DNA/RNA hybrid binding / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / regulation of type I interferon production / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Polymerase switching / Processive synthesis on the lagging strand ...DNA primase AEP / ribonucleotide binding / DNA replication initiation / DNA/RNA hybrid binding / Inhibition of replication initiation of damaged DNA by RB1/E2F1 / regulation of type I interferon production / Telomere C-strand synthesis initiation / alpha DNA polymerase:primase complex / Polymerase switching / Processive synthesis on the lagging strand / Removal of the Flap Intermediate / lagging strand elongation / mitotic DNA replication initiation / DNA replication, synthesis of primer / Polymerase switching on the C-strand of the telomere / DNA strand elongation involved in DNA replication / DNA synthesis involved in DNA repair / G1/S-Specific Transcription / leading strand elongation / DNA replication origin binding / DNA replication initiation / Activation of the pre-replicative complex / Defective pyroptosis / double-strand break repair via nonhomologous end joining / nuclear matrix / protein import into nucleus / DNA-directed RNA polymerase activity / nuclear envelope / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / ciliary basal body / DNA repair / nucleotide binding / intracellular membrane-bounded organelle / chromatin binding / protein kinase binding / chromatin / nucleolus / magnesium ion binding / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 7.41 Å | ||||||||||||
 Authors Authors | Cordoba JJ / Chazin WJ | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Flexibility and Distributive Synthesis Regulate RNA Priming and Handoff in Human DNA Polymerase α-Primase. Authors: John J Cordoba / Elwood A Mullins / Lauren E Salay / Brandt F Eichman / Walter J Chazin /  Abstract: DNA replication in eukaryotes relies on the synthesis of a ∼30-nucleotide RNA/DNA primer strand through the dual action of the heterotetrameric polymerase α-primase (pol-prim) enzyme. Synthesis of ...DNA replication in eukaryotes relies on the synthesis of a ∼30-nucleotide RNA/DNA primer strand through the dual action of the heterotetrameric polymerase α-primase (pol-prim) enzyme. Synthesis of the 7-10-nucleotide RNA primer is regulated by the C-terminal domain of the primase regulatory subunit (PRIM2C) and is followed by intramolecular handoff of the primer to pol α for extension by ∼20 nucleotides of DNA. Here, we provide evidence that RNA primer synthesis is governed by a combination of the high affinity and flexible linkage of the PRIM2C domain and the surprisingly low affinity of the primase catalytic domain (PRIM1) for substrate. Using a combination of small angle X-ray scattering and electron microscopy, we found significant variability in the organization of PRIM2C and PRIM1 in the absence and presence of substrate, and that the population of structures with both PRIM2C and PRIM1 in a configuration aligned for synthesis is low. Crosslinking was used to visualize the orientation of PRIM2C and PRIM1 when engaged by substrate as observed by electron microscopy. Microscale thermophoresis was used to measure substrate affinities for a series of pol-prim constructs, which showed that the PRIM1 catalytic domain does not bind the template or emergent RNA-primed templates with appreciable affinity. Together, these findings support a model of RNA primer synthesis in which generation of the nascent RNA strand and handoff of the RNA-primed template from primase to polymerase α is mediated by the high degree of inter-domain flexibility of pol-prim, the ready dissociation of PRIM1 from its substrate, and the much higher affinity of the POLA1cat domain of polymerase α for full-length RNA-primed templates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42035.map.gz emd_42035.map.gz | 31.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42035-v30.xml emd-42035-v30.xml emd-42035.xml emd-42035.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42035_fsc.xml emd_42035_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42035.png emd_42035.png | 26.3 KB | ||
| Filedesc metadata |  emd-42035.cif.gz emd-42035.cif.gz | 6.3 KB | ||
| Others |  emd_42035_half_map_1.map.gz emd_42035_half_map_1.map.gz emd_42035_half_map_2.map.gz emd_42035_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42035 http://ftp.pdbj.org/pub/emdb/structures/EMD-42035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42035 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42035 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42035.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42035.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.79015 Å | ||||||||||||||||||||||||||||||||||||
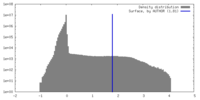
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42035_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
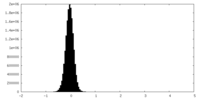
| Density Histograms |
-Half map: #2
| File | emd_42035_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of polymerase alpha-primase bound to an RNA prime...
| Entire | Name: Ternary complex of polymerase alpha-primase bound to an RNA primed-template substrate. |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of polymerase alpha-primase bound to an RNA prime...
| Supramolecule | Name: Ternary complex of polymerase alpha-primase bound to an RNA primed-template substrate. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Complex stabilized by BS3 crosslinking. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: DNA polymerase alpha subunit B
| Macromolecule | Name: DNA polymerase alpha subunit B / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GSATPSQKYN SRSNRGEVVT SFGLAQGV S WSGRGGAGNI SLKVLGCPEA LTGSYKSMFQ KLPDIREVLT CKIEELGSEL KEHYKIEAF TPLLAPAQEP VTLLGQIGCD SNGKLNNKSV ILEGDREHSS GAQIPVDLSE LKEYSLFPGQ VVIMEGINT TGRKLVATKL ...String: GSATPSQKYN SRSNRGEVVT SFGLAQGV S WSGRGGAGNI SLKVLGCPEA LTGSYKSMFQ KLPDIREVLT CKIEELGSEL KEHYKIEAF TPLLAPAQEP VTLLGQIGCD SNGKLNNKSV ILEGDREHSS GAQIPVDLSE LKEYSLFPGQ VVIMEGINT TGRKLVATKL YEGVPLPFYQ PTEEDADFEQ SMVLVACGPY TTSDSITYDP L LDLIAVIN HDRPDVCILF GPFLDAKHEQ VENCLLTSPF EDIFKQCLRT IIEGTRSSGS HL VFVPSLR DVHHEPVYPQ PPFSYSDLSR EDKKQVQFVS EPCSLSINGV IFGLTSTDLL FHL GAEEIS SSSGTSDRFS RILKHILTQR SYYPLYPPQE DMAIDYESFY VYAQLPVTPD VLII PSELR YFVKDVLGCV CVNPGRLTKG QVGGTFARLY LRRPAADGAE RQSPCIAVQV VRI UniProtKB: DNA polymerase alpha subunit B |
-Macromolecule #2: DNA polymerase alpha catalytic subunit
| Macromolecule | Name: DNA polymerase alpha catalytic subunit / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MAQLTDEEKY RDCERFKCPC PTCGTENIYD NVFDGSGTDM EPSLYRCSNI DCKASPL TF TVQLSNKLIM DIRRFIKKYY DGWLICEEPT CRNRTRHLPL QFSRTGPLCP ACMKATLQ P EYSDKSLYTQ LCFYRYIFDA ECALEKLTTD HEKDKLKKQF FTPKVLQDYR ...String: MAQLTDEEKY RDCERFKCPC PTCGTENIYD NVFDGSGTDM EPSLYRCSNI DCKASPL TF TVQLSNKLIM DIRRFIKKYY DGWLICEEPT CRNRTRHLPL QFSRTGPLCP ACMKATLQ P EYSDKSLYTQ LCFYRYIFDA ECALEKLTTD HEKDKLKKQF FTPKVLQDYR KLKNTAEQF LSRSGYSEVN LSKLFAGCAV KS UniProtKB: DNA polymerase alpha catalytic subunit |
-Macromolecule #3: DNA primase large subunit
| Macromolecule | Name: DNA primase large subunit / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQ YQSKLESELR KLKFSYRENL EDEYEPRRRD HISHFILRLA YCQSEELRRW F IQQEMDLL RFRFSILPKD KIQDFLKDSQ LQFEAISDEE KTLREQEIVA ...String: MEFSGRKWRK LRLAGDQRNA SYPHCLQFYL QPPSENISLI EFENLAIDRV KLLKSVENLG VSYVKGTEQ YQSKLESELR KLKFSYRENL EDEYEPRRRD HISHFILRLA YCQSEELRRW F IQQEMDLL RFRFSILPKD KIQDFLKDSQ LQFEAISDEE KTLREQEIVA SSPSLSGLKL GF ESIYKIP FADALDLFRG RKVYLEDGFA YVPLKDIVAI ILNEFRAKLS KALALTARSL PAV QSDERL QPLLNHLSHS YTGQDYSTQG NVGKISLDQI DLLSTKSFPP CMRQLHKALR ENHH LRHGG RMQYGLFLKG IGLTLEQALQ FWKQEFIKGK MDPDKFDKGY SYNIRHSFGK EGKRT DYTP FSCLKIILSN PPSQGDYHGC PFRHSDPELL KQKLQSYKIS PGGISQILDL VKGTHY QVA CQKYFEMIHN VDDCGFSLNH PNQFFCESQR ILNGGKDIKK EPIQPETPQP KPSVQKT KD ASSALASLNS SLEMDMEGLE DYFSEDS UniProtKB: DNA primase large subunit |
-Macromolecule #4: DNA primase small subunit
| Macromolecule | Name: DNA primase small subunit / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: GSMETFDPTE LPELLKLYYR RLFPYSQYYR WLNYGGVIKN YFQHREFSFT LKDDIYIRYQ SF NNQSDLE KEMQKMNPYK IDIGAVYSHR PNQHNTVKLG AFQAQEKELV FDIDMTDYDD VRR CCSSAD ICPKCWTLMT MAIRIIDRAL KEDFGFKHRL WVYSGRRGVH ...String: GSMETFDPTE LPELLKLYYR RLFPYSQYYR WLNYGGVIKN YFQHREFSFT LKDDIYIRYQ SF NNQSDLE KEMQKMNPYK IDIGAVYSHR PNQHNTVKLG AFQAQEKELV FDIDMTDYDD VRR CCSSAD ICPKCWTLMT MAIRIIDRAL KEDFGFKHRL WVYSGRRGVH CWVCDESVRK LSSA VRSGI VEYLSLVKGG QDVKKKVHLS EKIHPFIRKS INIIKKYFEE YALVNQDILE NKESW DKIL ALVPETIHDE LQQSFQKSHN SLQRWEHLKK VASRYQNNIK NDKYGPWLEW EIMLQY CFP RLDINVSKGI NHLLKSPFSV HPKTGRISVP IDLQKVDQFD PFTVPTISFI CRELDAI ST NEEEKEENEA ESDVKHRTRD YKKTSLAPYV KVFEHFLENL DKSRKGELLK KSDLQKDF UniProtKB: DNA primase small subunit |
-Macromolecule #5: RNA priming substrate - primer
| Macromolecule | Name: RNA priming substrate - primer / type: rna / ID: 5 / Details: 5' triphosphorylated |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGAUACUG |
-Macromolecule #6: RNA priming substrate - template
| Macromolecule | Name: RNA priming substrate - template / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GTATGTATGT CAGTATCCTG TATGTATGA |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.009 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: 2.5 uL sample was applied to grid for 1 minute, then blotted with filter paper, washed for 5 seconds with DI water, blotted, washed for 5 seconds in stain, blotted, then stained for 90 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1083 / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 1.2 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)