[English] 日本語
 Yorodumi
Yorodumi- EMDB-42142: Complete DNA termination subcomplex 2 of Xenopus laevis DNA polym... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||
 Map data Map data | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Primase / DNA polymerase / chimeric RNA-DNA primer / RNA/DNA hybrid / DNA replication / DNA synthesis / REPLICATION / TRANSFERASE-DNA-RNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationalpha DNA polymerase:primase complex / lagging strand elongation / mitotic DNA replication initiation / DNA replication, synthesis of primer / DNA strand elongation involved in DNA replication / leading strand elongation / DNA replication origin binding / DNA replication initiation / double-strand break repair via nonhomologous end joining / nuclear matrix ...alpha DNA polymerase:primase complex / lagging strand elongation / mitotic DNA replication initiation / DNA replication, synthesis of primer / DNA strand elongation involved in DNA replication / leading strand elongation / DNA replication origin binding / DNA replication initiation / double-strand break repair via nonhomologous end joining / nuclear matrix / nuclear envelope / single-stranded DNA binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / chromatin binding / chromatin / nucleolus / DNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | ||||||||||||
| Biological species | |||||||||||||
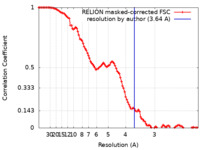
| Method | single particle reconstruction / cryo EM / Resolution: 3.64 Å | ||||||||||||
 Authors Authors | Mullins EA / Chazin WC / Eichman BF | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: A mechanistic model of primer synthesis from catalytic structures of DNA polymerase α-primase. Authors: Elwood A Mullins / Lauren E Salay / Clarissa L Durie / Noah P Bradley / Jane E Jackman / Melanie D Ohi / Walter J Chazin / Brandt F Eichman Abstract: The mechanism by which polymerase α-primase (polα-primase) synthesizes chimeric RNA-DNA primers of defined length and composition, necessary for replication fidelity and genome stability, is ...The mechanism by which polymerase α-primase (polα-primase) synthesizes chimeric RNA-DNA primers of defined length and composition, necessary for replication fidelity and genome stability, is unknown. Here, we report cryo-EM structures of polα-primase in complex with primed templates representing various stages of DNA synthesis. Our data show how interaction of the primase regulatory subunit with the primer 5'-end facilitates handoff of the primer to polα and increases polα processivity, thereby regulating both RNA and DNA composition. The structures detail how flexibility within the heterotetramer enables synthesis across two active sites and provide evidence that termination of DNA synthesis is facilitated by reduction of polα and primase affinities for the varied conformations along the chimeric primer/template duplex. Together, these findings elucidate a critical catalytic step in replication initiation and provide a comprehensive model for primer synthesis by polα-primase. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42142.map.gz emd_42142.map.gz | 80.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42142-v30.xml emd-42142-v30.xml emd-42142.xml emd-42142.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42142_fsc.xml emd_42142_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42142.png emd_42142.png | 27.6 KB | ||
| Filedesc metadata |  emd-42142.cif.gz emd-42142.cif.gz | 7.4 KB | ||
| Others |  emd_42142_additional_1.map.gz emd_42142_additional_1.map.gz emd_42142_half_map_1.map.gz emd_42142_half_map_1.map.gz emd_42142_half_map_2.map.gz emd_42142_half_map_2.map.gz | 92.6 MB 80.9 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42142 http://ftp.pdbj.org/pub/emdb/structures/EMD-42142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42142 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42142 | HTTPS FTP |
-Related structure data
| Related structure data |  8ucwMC  8g99C  8g9fC  8g9lC  8g9nC  8g9oC  8ucuC  8ucvC  8v5mC  8v5nC  8v5oC  8v6gC  8v6hC  8v6iC  8v6jC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42142.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42142.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09333 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Complete DNA termination subcomplex 2 of Xenopus laevis...
| File | emd_42142_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase (locally sharpened) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Complete DNA termination subcomplex 2 of Xenopus laevis...
| File | emd_42142_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase (half 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Complete DNA termination subcomplex 2 of Xenopus laevis...
| File | emd_42142_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase (half 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Polymerase alpha-primase with a DNA termination substrate
| Entire | Name: Polymerase alpha-primase with a DNA termination substrate |
|---|---|
| Components |
|
-Supramolecule #1: Polymerase alpha-primase with a DNA termination substrate
| Supramolecule | Name: Polymerase alpha-primase with a DNA termination substrate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Heterotetrameric protein complex with an RNA-DNA/DNA duplex |
|---|---|
| Molecular weight | Theoretical: 330 KDa |
-Supramolecule #2: Polymerase alpha
| Supramolecule | Name: Polymerase alpha / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: Heterodimer consisting of POLA1 and POLA2 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 200 KDa |
-Supramolecule #3: Primase
| Supramolecule | Name: Primase / type: complex / ID: 3 / Parent: 1 / Details: Heterodimer consisting of PRIM1 and PRIM2 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 110 KDa |
-Supramolecule #4: DNA termination substrate
| Supramolecule | Name: DNA termination substrate / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2-#3 / Details: RNA-DNA/DNA duplex |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 30 KDa |
-Macromolecule #1: DNA polymerase alpha catalytic subunit
| Macromolecule | Name: DNA polymerase alpha catalytic subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 129.009414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAADGSQVF RFYWLDAYED QYSQPGVVYL FGKVWIESAD AYVSCCVSVK NIERTVYLLP RENRVQLSTG KDTGAPVSMM HVYQEFNEA VAEKYKIMKF KSKKVDKDYA FEIPDVPASS EYLEVRYSAD SPQLPQDLKG ETFSHVFGTN TSSLELFLLS R KIKGPSWL ...String: SNAADGSQVF RFYWLDAYED QYSQPGVVYL FGKVWIESAD AYVSCCVSVK NIERTVYLLP RENRVQLSTG KDTGAPVSMM HVYQEFNEA VAEKYKIMKF KSKKVDKDYA FEIPDVPASS EYLEVRYSAD SPQLPQDLKG ETFSHVFGTN TSSLELFLLS R KIKGPSWL EIKSPQLSSQ PMSWCKVEAV VTRPDQVSVV KDLAPPPVVV LSLSMKTVQN AKTHQNEIVA IAALVHHTFP LD KAPPQPP FQTHFCVLSK LNDCIFPYDY NEAVKQKNAN IEIALTERTL LGFFLAKIHK IDPDVIVGHD IYGFDLEVLL QRI NSCKVP FWSKIGRLRR SVMPKLGGRS GFAERNAACG RIICDIEISA KELIRCKSYH LSELVHQILK AERVVIPPEN IRNA YNDSV HLLYMLENTW IDAKFILQIM CELNVLPLAL QITNIAGNVM SRTLMGGRSE RNEYLLLHAF TENNFIVPDK PVFKK MQQT TVEDNDDMGT DQNKNKSRKK AAYAGGLVLE PKVGFYDKFI LLLDFNSLYP SIIQEYNICF TTVHREAPST QKGEDQ DEI PELPHSDLEM GILPREIRKL VERRRHVKQL MKQPDLNPDL YLQYDIRQKA LKLTANSMYG CLGFSYSRFY AKPLAAL VT HQGREILLHT KEMVQKMNLE VIYGDTDSIM INTNCNNLEE VFKLGNRVKS EINKSYKLLE IDIDGIFKSL LLLKKKKY A ALTVEPTGDG KYVTKQELKG LDIVRRDWCE LAKQAGNYVI SQILSDQPRD SIVENIQKKL TEIGENVTNG TVPITQYEI NKALTKDPQD YPDKKSLPHV HVALWINSQG GRKVKAGDTI SYVICQDGSN LSASQRAYAQ EQLQKQENLS IDTQYYLSQQ VHPVVARIC EPIDGIDSAL IAMWLGLDPS QFRAHRHYQQ DEENDALLGG PSQLTDEEKY RDCERFKFFC PKCGTENIYD N VFDGSGLQ IEPGLKRCSK PECDASPLDY VIQVHNKLLL DIRRYIKKYY SGWLVCEEKT CQNRTRRLPL SFSRNGPICQ AC SKATLRS EYPEKALYTQ LCFYRFIFDW DYALEKVVSE QERGHLKKKL FQESENQYKK LKSTVDQVLS RSGYSEVNLS KLF QTLNTI K UniProtKB: DNA polymerase alpha catalytic subunit |
-Macromolecule #2: DNA template
| Macromolecule | Name: DNA template / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 18.203668 KDa |
| Sequence | String: (DT)(DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DG) (DT)(DA)(DT)(DG)(DT)(DC)(DG)(DC)(DT)(DA) (DC)(DA)(DA)(DT)(DC)(DG)(DC)(DT)(DA) (DA)(DG)(DT)(DT)(DC)(DA)(DC)(DG)(DC)(DA) (DG) (DT)(DA)(DT)(DC)(DC)(DT) ...String: (DT)(DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DG) (DT)(DA)(DT)(DG)(DT)(DC)(DG)(DC)(DT)(DA) (DC)(DA)(DA)(DT)(DC)(DG)(DC)(DT)(DA) (DA)(DG)(DT)(DT)(DC)(DA)(DC)(DG)(DC)(DA) (DG) (DT)(DA)(DT)(DC)(DC)(DT)(DG)(DT) (DA)(DT)(DG)(DT)(DA)(DT)(DG)(DT)(DA)(DT) (DG) |
-Macromolecule #3: RNA-DNA/DNA primer
| Macromolecule | Name: RNA-DNA/DNA primer / type: other / ID: 3 / Number of copies: 1 Classification: polydeoxyribonucleotide/polyribonucleotide hybrid |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.249697 KDa |
| Sequence | String: (GTP)GAUACUGC(DG) (DT)(DG)(DA)(DA)(DC)(DT)(DT)(DA)(DG)(DC) (DG)(DA)(DT)(DT)(DG) (DT)(DA)(DG)(DOC) |
-Macromolecule #4: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: DGT |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-DGT: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 22893 / Average electron dose: 55.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)