-Search query
-Search result
Showing 1 - 50 of 98 items for (author: basak & ak)

EMDB-35163: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

EMDB-35164: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state
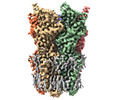
EMDB-36339: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5
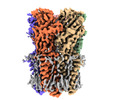
EMDB-37446: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state
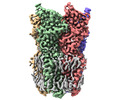
EMDB-37447: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

PDB-8i47: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

PDB-8i48: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state

PDB-8jj3: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5

PDB-8wcq: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state

PDB-8wcr: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

EMDB-35161: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

EMDB-35162: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

PDB-8i41: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

PDB-8i42: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

EMDB-36137: 
Cryo-EM structure of Holo form of ScBfr with O symmetry

EMDB-36139: 
Cryo-EM structure of Holo form of ScBfr in C1 symmetry

EMDB-36140: 
Cryo-EM structure of Fe-biomineral from bacterioferritin

PDB-8jax: 
Cryo-EM structure of Holo form of ScBfr with O symmetry

PDB-8jb0: 
Cryo-EM structure of Holo form of ScBfr in C1 symmetry

EMDB-33639: 
Cryo-EM structure of Apo form of ScBfr

EMDB-33640: 
Cryo-EM structure of bacterioferritin holoform 1a

EMDB-33645: 
Cryo-EM structure if bacterioferritin holoform

PDB-7y6f: 
Cryo-EM structure of Apo form of ScBfr

PDB-7y6g: 
Cryo-EM structure of bacterioferritin holoform 1a

PDB-7y6p: 
Cryo-EM structure if bacterioferritin holoform

EMDB-33614: 
Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase

EMDB-33615: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1)

EMDB-33616: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2)

EMDB-33617: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone)

PDB-7y5a: 
Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase

PDB-7y5b: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1)

PDB-7y5c: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2)

PDB-7y5d: 
Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone)

EMDB-23700: 
Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol

EMDB-23701: 
Full length alpha1 Glycine receptor in presence of 0.1mM Glycine

EMDB-23702: 
Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol

EMDB-23703: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine

EMDB-23704: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1

EMDB-23705: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 2

EMDB-23706: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 3

PDB-7m6m: 
Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol

PDB-7m6n: 
Full length alpha1 Glycine receptor in presence of 0.1mM Glycine

PDB-7m6o: 
Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol

PDB-7m6p: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine

PDB-7m6q: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1

PDB-7m6r: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 2

PDB-7m6s: 
Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 3

EMDB-32524: 
Cryo-EM map of E.coli FtsH AAA protease

EMDB-32520: 
Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex
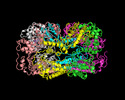
EMDB-32521: 
Cryo-EM structure of E.Coli FtsH protease cytosolic domains
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
