[English] 日本語
 Yorodumi
Yorodumi- EMDB-33617: Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegma... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / F-ATP synthase / cryo-EM / mycobacteria / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / hydrolase activity / lipid binding / ATP hydrolysis activity ...proton motive force-driven plasma membrane ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATPase activity, rotational mechanism / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / hydrolase activity / lipid binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) | |||||||||
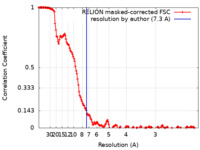
| Method | single particle reconstruction / cryo EM / Resolution: 7.3 Å | |||||||||
 Authors Authors | Saw W-G / Wong CF / Grueber G | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Antimicrob Agents Chemother / Year: 2022 Journal: Antimicrob Agents Chemother / Year: 2022Title: Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors. Authors: Chui Fann Wong / Wuan-Geok Saw / Sandip Basak / Mio Sano / Hiroshi Ueno / Hwee Wen Kerk / Dennis Litty / Priya Ragunathan / Thomas Dick / Volker Müller / Hiroyuki Noji / Gerhard Grüber /     Abstract: The FF-ATP synthase is required for the viability of tuberculosis (TB) and nontuberculous mycobacteria (NTM) and has been validated as a drug target. Here, we present the cryo-EM structures of the ...The FF-ATP synthase is required for the viability of tuberculosis (TB) and nontuberculous mycobacteria (NTM) and has been validated as a drug target. Here, we present the cryo-EM structures of the Mycobacterium smegmatis F-ATPase and the FF-ATP synthase with different nucleotide occupation within the catalytic sites and visualize critical elements for latent ATP hydrolysis and efficient ATP synthesis. Mutational studies reveal that the extended C-terminal domain (αCTD) of subunit α is the main element for the self-inhibition mechanism of ATP hydrolysis for TB and NTM bacteria. Rotational studies indicate that the transition between the inhibition state by the αCTD and the active state is a rapid process. We demonstrate that the unique mycobacterial γ-loop and subunit δ are critical elements required for ATP formation. The data underline that these mycobacterium-specific elements of α, γ, and δ are attractive targets, providing a platform for the discovery of species-specific inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33617.map.gz emd_33617.map.gz | 22.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33617-v30.xml emd-33617-v30.xml emd-33617.xml emd-33617.xml | 30.3 KB 30.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33617_fsc.xml emd_33617_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33617.png emd_33617.png | 128.6 KB | ||
| Masks |  emd_33617_msk_1.map emd_33617_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33617.cif.gz emd-33617.cif.gz | 8 KB | ||
| Others |  emd_33617_half_map_1.map.gz emd_33617_half_map_1.map.gz emd_33617_half_map_2.map.gz emd_33617_half_map_2.map.gz | 259.6 MB 259.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33617 http://ftp.pdbj.org/pub/emdb/structures/EMD-33617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33617 | HTTPS FTP |
-Validation report
| Summary document |  emd_33617_validation.pdf.gz emd_33617_validation.pdf.gz | 906.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33617_full_validation.pdf.gz emd_33617_full_validation.pdf.gz | 905.8 KB | Display | |
| Data in XML |  emd_33617_validation.xml.gz emd_33617_validation.xml.gz | 22.9 KB | Display | |
| Data in CIF |  emd_33617_validation.cif.gz emd_33617_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33617 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33617 | HTTPS FTP |
-Related structure data
| Related structure data |  7y5dMC  7y5aC  7y5bC  7y5cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33617.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33617.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
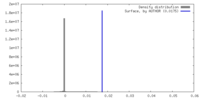
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33617_msk_1.map emd_33617_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
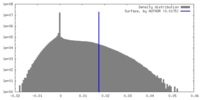
| Density Histograms |
-Half map: #2
| File | emd_33617_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
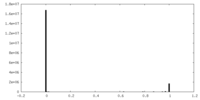
| Density Histograms |
-Half map: #1
| File | emd_33617_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
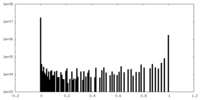
| Density Histograms |
- Sample components
Sample components
+Entire : F-ATP synthase
+Supramolecule #1: F-ATP synthase
+Macromolecule #1: ATP synthase subunit a
+Macromolecule #2: ATP synthase subunit b
+Macromolecule #3: ATP synthase subunit b-delta
+Macromolecule #4: ATP synthase subunit c
+Macromolecule #5: ATP synthase subunit alpha
+Macromolecule #6: ATP synthase subunit alpha
+Macromolecule #7: ATP synthase subunit beta
+Macromolecule #8: ATP synthase gamma chain
+Macromolecule #9: ATP synthase epsilon chain
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3663 / Average electron dose: 38.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)