-Search query
-Search result
Showing all 38 items for (author: randall & s)

EMDB-27781: 
Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide
Method: single particle / : Martin GM, Ward AB

EMDB-27784: 
Cryo-EM structure of 239 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27785: 
Cryo-EM structure of 311 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27786: 
Cryo-EM structure of 334 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27787: 
Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27788: 
Cryo-EM structure of 356 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27789: 
Cryo-EM structure of 364 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyt: 
Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide
Method: single particle / : Martin GM, Ward AB

PDB-8dyw: 
Cryo-EM structure of 239 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyx: 
Cryo-EM structure of 311 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyy: 
Cryo-EM structure of 334 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz3: 
Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz4: 
Cryo-EM structure of 356 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz5: 
Cryo-EM structure of 364 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-26715: 
Structure of HIV-1 capsid declination
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT
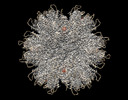
EMDB-26718: 
T=1 particle HIV-1 CA G60A/G61P/M66A
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT
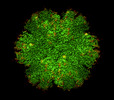
EMDB-28054: 
T=1 particle HIV-1 CA G60A/G61P
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

EMDB-28057: 
T=1 particle HIV-1 CA M66A
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

EMDB-28186: 
Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT, dos Santos NFB

PDB-7urn: 
Structure of HIV-1 capsid declination
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

PDB-7urt: 
T=1 particle HIV-1 CA G60A/G61P/M66A
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

PDB-8eep: 
T=1 particle HIV-1 CA G60A/G61P
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

PDB-8eet: 
T=1 particle HIV-1 CA M66A
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT

PDB-8ejl: 
Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide
Method: single particle / : Pornillos O, Ganser-Pornillos BK, Schirra RT, dos Santos NFB

EMDB-11953: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-11954: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14810: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14811: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-24614: 
Cryo-EM structure of murine Dispatched 'R' conformation
Method: single particle / : Asarnow D, Wang Q

EMDB-24615: 
Cryo-EM structure of murine Dispatched 'T' conformation
Method: single particle / : Asarnow D, Wang Q

EMDB-24616: 
Cryo-EM structure of murine Dispatched NNN mutant
Method: single particle / : Asarnow D, Wang Q, Ding K, Cheng Y, Beachy PA

EMDB-24617: 
Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog
Method: single particle / : Asarnow D, Wang Q

PDB-7rph: 
Cryo-EM structure of murine Dispatched 'R' conformation
Method: single particle / : Asarnow D, Wang Q, Ding K, Cheng Y, Beachy PA

PDB-7rpi: 
Cryo-EM structure of murine Dispatched 'T' conformation
Method: single particle / : Asarnow D, Wang Q, Ding K, Cheng Y, Beachy PA

PDB-7rpj: 
Cryo-EM structure of murine Dispatched NNN mutant
Method: single particle / : Asarnow D, Wang Q, Ding K, Cheng Y, Beachy PA

PDB-7rpk: 
Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog
Method: single particle / : Asarnow D, Wang Q, Ding K, Cheng Y, Beachy PA

EMDB-8582: 
Structure of the HIV-1 Capsid Protein and spacer peptide 1 by Cryo-EM
Method: helical / : Zhang P, Randall S

PDB-5up4: 
Structure of the HIV-1 Capsid Protein and spacer peptide 1 by Cryo-EM
Method: helical / : Perilla JR, Schirra R, Zhang P, Schulten K
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
