+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24615 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of murine Dispatched 'T' conformation | ||||||||||||||||||
 Map data Map data | DISP1-A "T" - auto-sharpened map from cryoSPARC nonuniform refinement (paper version), units of standard deviation above mean density. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | RND transporter / Hedgehog binding / Sterol sensing domain / sodium binding / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpatched ligand maturation / diaphragm development / embryonic pattern specification / dorsal/ventral pattern formation / peptide transport / determination of left/right symmetry / molecular carrier activity / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||||||||
 Authors Authors | Asarnow D / Wang Q | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Dispatched uses Na flux to power release of lipid-modified Hedgehog. Authors: Qianqian Wang / Daniel E Asarnow / Ke Ding / Randall K Mann / Jason Hatakeyama / Yunxiao Zhang / Yong Ma / Yifan Cheng / Philip A Beachy /   Abstract: The Dispatched protein, which is related to the NPC1 and PTCH1 cholesterol transporters and to H-driven transporters of the RND family, enables tissue-patterning activity of the lipid-modified ...The Dispatched protein, which is related to the NPC1 and PTCH1 cholesterol transporters and to H-driven transporters of the RND family, enables tissue-patterning activity of the lipid-modified Hedgehog protein by releasing it from tightly -localized sites of embryonic expression. Here we determine a cryo-electron microscopy structure of the mouse protein Dispatched homologue 1 (DISP1), revealing three Na ions coordinated within a channel that traverses its transmembrane domain. We find that the rate of Hedgehog export is dependent on the Na gradient across the plasma membrane. The transmembrane channel and Na binding are disrupted in DISP1-NNN, a variant with asparagine substitutions for three intramembrane aspartate residues that each coordinate and neutralize the charge of one of the three Na ions. DISP1-NNN and variants that disrupt single Na sites retain binding to, but are impaired in export of the lipid-modified Hedgehog protein to the SCUBE2 acceptor. Interaction of the amino-terminal signalling domain of the Sonic hedgehog protein (ShhN) with DISP1 occurs via an extensive buried surface area and contacts with an extended furin-cleaved DISP1 arm. Variability analysis reveals that ShhN binding is restricted to one extreme of a continuous series of DISP1 conformations. The bound and unbound DISP1 conformations display distinct Na-site occupancies, which suggests a mechanism by which transmembrane Na flux may power extraction of the lipid-linked Hedgehog signal from the membrane. Na-coordinating residues in DISP1 are conserved in PTCH1 and other metazoan RND family members, suggesting that Na flux powers their conformationally driven activities. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24615.map.gz emd_24615.map.gz | 114.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24615-v30.xml emd-24615-v30.xml emd-24615.xml emd-24615.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24615.png emd_24615.png | 132.7 KB | ||
| Masks |  emd_24615_msk_1.map emd_24615_msk_1.map emd_24615_msk_2.map emd_24615_msk_2.map | 125 MB 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24615.cif.gz emd-24615.cif.gz | 7.6 KB | ||
| Others |  emd_24615_additional_1.map.gz emd_24615_additional_1.map.gz emd_24615_additional_2.map.gz emd_24615_additional_2.map.gz emd_24615_half_map_1.map.gz emd_24615_half_map_1.map.gz emd_24615_half_map_2.map.gz emd_24615_half_map_2.map.gz | 118.1 MB 62.8 MB 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24615 http://ftp.pdbj.org/pub/emdb/structures/EMD-24615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24615 | HTTPS FTP |
-Related structure data
| Related structure data |  7rpiMC  7rphC  7rpjC  7rpkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24615.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24615.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DISP1-A "T" - auto-sharpened map from cryoSPARC nonuniform refinement (paper version), units of standard deviation above mean density. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24615_msk_1.map emd_24615_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
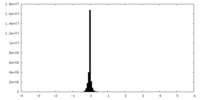
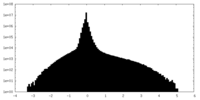
| Density Histograms |
-Mask #2
| File |  emd_24615_msk_2.map emd_24615_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
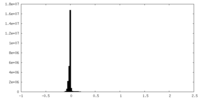
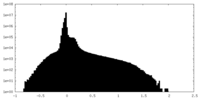
| Density Histograms |
-Additional map: DISP1-A "T" - auto-sharpened map from cryoSPARC nonuniform...
| File | emd_24615_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DISP1-A "T" - auto-sharpened map from cryoSPARC nonuniform refinement (paper version). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DISP1-A "T" - raw map from cryoSPARC nonuniform...
| File | emd_24615_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DISP1-A "T" - raw map from cryoSPARC nonuniform refinement (paper version). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: DISP1-A "T" - half map 1 from cryoSPARC...
| File | emd_24615_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DISP1-A "T" - half map 1 from cryoSPARC nonuniform refinement (paper version). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: DISP1-A "T" - half map 2 from cryoSPARC...
| File | emd_24615_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DISP1-A "T" - half map 2 from cryoSPARC nonuniform refinement (paper version). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dispatched protein 'T' conformation
| Entire | Name: Dispatched protein 'T' conformation |
|---|---|
| Components |
|
-Supramolecule #1: Dispatched protein 'T' conformation
| Supramolecule | Name: Dispatched protein 'T' conformation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.91493 KDa |
-Macromolecule #1: Protein dispatched homolog 1
| Macromolecule | Name: Protein dispatched homolog 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 152.071141 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARPFKFPRS YAALLADWPV VVLGMCTLLI VVCALVGVLV PELPDFSDPL LGFEPRGTTI GQRLVTWNNM MRNTGYKATL ANYPYKYAE EQARSHRDDR WSDDHHERER REVDWNFQKD SFFCDVPSDG YSRVVFASAG GETLWNLPAI KSMCDVDNSR I RSHPQFSD ...String: MARPFKFPRS YAALLADWPV VVLGMCTLLI VVCALVGVLV PELPDFSDPL LGFEPRGTTI GQRLVTWNNM MRNTGYKATL ANYPYKYAE EQARSHRDDR WSDDHHERER REVDWNFQKD SFFCDVPSDG YSRVVFASAG GETLWNLPAI KSMCDVDNSR I RSHPQFSD LCQRTTAVSC CPSWTLGNYI AILNNRSSCQ KIVERDVSHT LKLLRTCAKH YQNGTLGPDC WDKAARRKDQ LK CTNVPRK CTKYNAVYQI LHYLVDKDFM TPKTADYAVP ALKYSMLFSP TEKGESMMNI YLDNFENWNS SDGITTVTGI EFG IKHSLF QDYLLMDTVY PAIAIAIVLL IMCVYTKSMF ITLMTMFAII SSLIVSYFLY RVVFNFEFFP FMNLTALIIL VGIG ADDAF VLCDVWNYTK FDKPRAETSE AVSVTLQHAA LSMFVTSFTT AAAFYANYVS NITAIRCFGV YAGTAILVNY VLMVT WLPA VIVLHERYLL NIFTCFRKPQ PQAYDKSCWA VLCQKCRRVL FAVSEASRIF FEKVLPCIVI KFRYLWLIWF LALTVG GAY IVCVNPKMKL PSLELSEFQV FRSSHPFERY DAEFKKLFMF ERVHHGEELH MPITVIWGVS PEDSGDPLNP KSKGELT LD STFNIASPAS QAWILHFCQK LRNQTFFHQT EQQDFTSCFI ETFKQWMENQ DCDEPALYPC CSHCSFPYKQ EVFELCIK K AIMELDRSTG YHLNNKTPGP RFDINDTIRA VVLEFQSTFL FTLAYEKMQQ FYKEVDSWIS HELSSAPEGL SRGWFVSNL EFYDLQDSLS DGTLIAMGLS VAVAFSVMLL TTWNIIISLY AIVSIAGTIF VTVGSLVLLG WELNVLESVT ISVAVGLSVD FAVHYGVAY RLAPDPDREG KVIFSLSRMG SAIAMAALTT FVAGAMMMPS TVLAYTQLGT FMMLVMCVSW AFATFFFQCL C RCLGPQGT CGQIPFPTKL QCSPFSHTLS ARPGDRGPSK THAASAYSVD ARGQKSQLEH EFYELQPLAS HSCTSSEKTT YE EPHTCSE FFNGQAKNLR MPVPAAYSSE LTKSPSSEPG SALLQSCLEQ DTVCHFSLNP RCNCRDAYTH LQYGLPEIHC QQM GDSLCH KCASTAGGFV QIQSSVAPLK ASHQAAEGLL HPAQHMLPPG MQNSRPRNFF LHSVQHFQAQ ENLGRTSTHS TDER LPRTA ELSPPPSDSR STESFQRACC HPENNQRRLC KSRDPGDTEG SGGTKSKVSG LPNQTDKEEK QVEPSLLQTD ETVNS EHLN HNESNFTFSH LPGEAGCRSC PNSPQSCRSI MRSKCGTEDC QTPNLEANVP AVPTHSDLSG ESLLIKTL UniProtKB: Protein dispatched homolog 1 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 26 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 3 / Number of copies: 2 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 5 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 43 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.56 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV Details: Wait time 20 seconds, blot time 4 seconds, blotting force 0. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Automated data collection in SerialEM using 3x3 image shift pattern (one shot per hole), using beam tilt compensation. |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4487 / Average electron dose: 66.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 59880 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 72.1 |
|---|---|
| Output model |  PDB-7rpi: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)