+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T=1 particle HIV-1 CA G60A/G61P/M66A | |||||||||
 Map data Map data | T=1 particle of HIV-1 CA G60A/G61P/M66A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / capsid / declination / pentamer / hexamer / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost multivesicular body / viral nucleocapsid / viral translational frameshifting / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
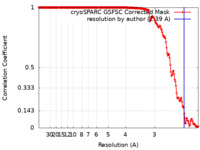
| Method | single particle reconstruction / cryo EM / Resolution: 2.39 Å | |||||||||
 Authors Authors | Pornillos O / Ganser-Pornillos BK / Schirra RT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: A molecular switch modulates assembly and host factor binding of the HIV-1 capsid. Authors: Randall T Schirra / Nayara F B Dos Santos / Kaneil K Zadrozny / Iga Kucharska / Barbie K Ganser-Pornillos / Owen Pornillos /   Abstract: The HIV-1 capsid is a fullerene cone made of quasi-equivalent hexamers and pentamers of the viral CA protein. Typically, quasi-equivalent assembly of viral capsid subunits is controlled by a ...The HIV-1 capsid is a fullerene cone made of quasi-equivalent hexamers and pentamers of the viral CA protein. Typically, quasi-equivalent assembly of viral capsid subunits is controlled by a molecular switch. Here, we identify a Thr-Val-Gly-Gly motif that modulates CA hexamer/pentamer switching by folding into a 3 helix in the pentamer and random coil in the hexamer. Manipulating the coil/helix configuration of the motif allowed us to control pentamer and hexamer formation in a predictable manner, thus proving its function as a molecular switch. Importantly, the switch also remodels the common binding site for host factors that are critical for viral replication and the new ultra-potent HIV-1 inhibitor lenacapavir. This study reveals that a critical assembly element also modulates the post-assembly and viral replication functions of the HIV-1 capsid and provides new insights on capsid function and inhibition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26718.map.gz emd_26718.map.gz | 484.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26718-v30.xml emd-26718-v30.xml emd-26718.xml emd-26718.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26718_fsc.xml emd_26718_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26718.png emd_26718.png | 143.6 KB | ||
| Filedesc metadata |  emd-26718.cif.gz emd-26718.cif.gz | 5.5 KB | ||
| Others |  emd_26718_half_map_1.map.gz emd_26718_half_map_1.map.gz emd_26718_half_map_2.map.gz emd_26718_half_map_2.map.gz | 474.7 MB 474.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26718 http://ftp.pdbj.org/pub/emdb/structures/EMD-26718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26718 | HTTPS FTP |
-Validation report
| Summary document |  emd_26718_validation.pdf.gz emd_26718_validation.pdf.gz | 856.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26718_full_validation.pdf.gz emd_26718_full_validation.pdf.gz | 856.3 KB | Display | |
| Data in XML |  emd_26718_validation.xml.gz emd_26718_validation.xml.gz | 26.3 KB | Display | |
| Data in CIF |  emd_26718_validation.cif.gz emd_26718_validation.cif.gz | 34.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26718 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26718 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26718 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26718 | HTTPS FTP |
-Related structure data
| Related structure data |  7urtMC  7urnC  8eepC  8eetC  8ejlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26718.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26718.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T=1 particle of HIV-1 CA G60A/G61P/M66A | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_26718_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
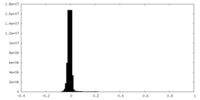
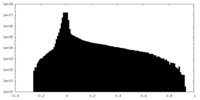
| Density Histograms |
-Half map: Half map B
| File | emd_26718_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
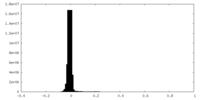
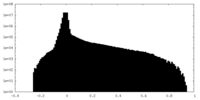
| Density Histograms |
- Sample components
Sample components
-Entire : T=1 particle of HIV-1 CA G60A/G61P/M66A
| Entire | Name: T=1 particle of HIV-1 CA G60A/G61P/M66A |
|---|---|
| Components |
|
-Supramolecule #1: T=1 particle of HIV-1 CA G60A/G61P/M66A
| Supramolecule | Name: T=1 particle of HIV-1 CA G60A/G61P/M66A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Gag polyprotein
| Macromolecule | Name: Gag polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 25.624396 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PIVQNLQGQM VHQAISPRTL NAWVKVVEEK AFSPEVIPMF SALSEGATPQ DLNTMLNTVA PHQAAAQMLK ETINEEAAEW DRLHPVHAG PIAPGQMREP RGSDIAGTTS TLQEQIGWMT HNPPIPVGEI YKRWIILGLN KIVRMYSPTS ILDIRQGPKE P FRDYVDRF ...String: PIVQNLQGQM VHQAISPRTL NAWVKVVEEK AFSPEVIPMF SALSEGATPQ DLNTMLNTVA PHQAAAQMLK ETINEEAAEW DRLHPVHAG PIAPGQMREP RGSDIAGTTS TLQEQIGWMT HNPPIPVGEI YKRWIILGLN KIVRMYSPTS ILDIRQGPKE P FRDYVDRF YKTLRAEQAS QEVKNWMTET LLVQNANPDC KTILKALGPG ATLEEMMTAC QGVGGPGHKA RVL UniProtKB: Gag polyprotein |
-Macromolecule #2: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Manual plunge-freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



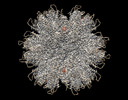





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)