-Search query
-Search result
Showing 1 - 50 of 113 items for (author: macher & m)

EMDB-41228: 
Cryo-EM structure of the Methanosarcina mazei apo glutamin synthetase structure: dodecameric form
Method: single particle / : Schumacher MA

EMDB-41229: 
Cryo-EM structure of Methanosarcina mazie glutamine synthetase captured as partial oligomer
Method: single particle / : Schumacher MA

EMDB-41232: 
Cryo-EM structure of the Methanosarcina mazei glutamine synthetase (GS) with Met-Sox-P and ADP
Method: single particle / : Schumacher MA

PDB-8tfb: 
Cryo-EM structure of the Methanosarcina mazei apo glutamin synthetase structure: dodecameric form
Method: single particle / : Schumacher MA

PDB-8tfc: 
Cryo-EM structure of Methanosarcina mazie glutamine synthetase captured as partial oligomer
Method: single particle / : Schumacher MA

PDB-8tfk: 
Cryo-EM structure of the Methanosarcina mazei glutamine synthetase (GS) with Met-Sox-P and ADP
Method: single particle / : Schumacher MA
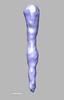
EMDB-28834: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 3
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG
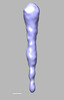
EMDB-28838: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 2
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG

EMDB-28839: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 1
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG
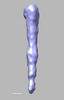
EMDB-28840: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 4
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG
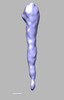
EMDB-28841: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 5
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG
EMDB-28842: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 6
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG

EMDB-28843: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 7
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG

EMDB-28844: 
EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 8
Method: subtomogram averaging / : Ruiz T, Radermacher M, Mintz KP, Tang-Siegel GG

EMDB-27460: 
HMGCR-UBIAD1 Complex Dimer
Method: single particle / : Chen H, Qi X, Li X

EMDB-27461: 
HMGCR-UBIAD1 Complex Monomer
Method: single particle / : Chen H, Qi X, Li X

EMDB-27475: 
HMGCR-UBIAD1-BRIL-Fab-Nb Complex
Method: single particle / : Chen H, Qi X, Li X

EMDB-27477: 
HMGCR(40-55 deletion)-UBIAD1 Complex Dimer
Method: single particle / : Chen H, Qi X, Li X

EMDB-27478: 
HMGCR(40-55 deletion)-UBIAD1 Complex Monomer
Method: single particle / : Chen H, Qi X, Li X

EMDB-26824: 
Citrus V-ATPase State 1, Highest-Resolution Class
Method: single particle / : Tan YZ, Rubinstein JL

EMDB-26825: 
Citrus V-ATPase State 1, H in contact with subunit a
Method: single particle / : Keon KA, Abdelaziz RA, Schulze WX, Schumacher K, Rubinstein JL

EMDB-26826: 
Citrus V-ATPase State 1, H in contact with subunits AB
Method: single particle / : Abdelaziz RA, Keon KA, Schulze WX, Schumacher K, Rubinstein JL

EMDB-26827: 
Citrus V-ATPase State 2, Highest-Resolution Class
Method: single particle / : Tan YZ, Rubinstein JL

EMDB-26828: 
Citrus V-ATPase State 2, H in contact with subunit a
Method: single particle / : Tan YZ, Rubinstein JL

EMDB-26829: 
Citrus V-ATPase State 2, H in contact with subunits AB
Method: single particle / : Tan YZ, Rubinstein JL

PDB-7uw9: 
Citrus V-ATPase State 1, H in contact with subunit a
Method: single particle / : Keon KA, Abdelaziz RA, Schulze WX, Schumacher K, Rubinstein JL

PDB-7uwa: 
Citrus V-ATPase State 1, H in contact with subunits AB
Method: single particle / : Abdelaziz RA, Keon KA, Schulze WX, Schumacher K, Rubinstein JL

PDB-7uwb: 
Citrus V-ATPase State 2, Highest-Resolution Class
Method: single particle / : Keon KA, Abdelaziz RA, Schulze WX, Schumacher K, Rubinstein JL

PDB-7uwc: 
Citrus V-ATPase State 2, H in contact with subunit a
Method: single particle / : Keon KA, Abdelaziz RA, Schulze WX, Schumacher K, Rubinstein JL

PDB-7uwd: 
Citrus V-ATPase State 2, H in contact with subunits AB
Method: single particle / : Keon KA, Abdelaziz RA, Schulze WX, Schumacher K, Rubinstein JL

EMDB-25863: 
S. aureus GS(12)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J

EMDB-25864: 
S. aureus GS(12) - apo
Method: single particle / : Travis BA, Peck J

EMDB-25866: 
L. monocytogenes GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J

EMDB-25867: 
P. polymyxa GS(12)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J

EMDB-25868: 
P. polymyxa GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J

EMDB-25869: 
B. subtilis GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J

EMDB-25870: 
P. polymyxa GS(12) - apo
Method: single particle / : Travis BA, Peck J

EMDB-25871: 
L. monocytogenes GS(12) - apo
Method: single particle / : Travis BA, Peck J

PDB-7tf6: 
S. aureus GS(12)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tf7: 
S. aureus GS(12) - apo
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tf9: 
L. monocytogenes GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tfa: 
P. polymyxa GS(12)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tfb: 
P. polymyxa GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tfc: 
B. subtilis GS(14)-Q-GlnR peptide
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tfd: 
P. polymyxa GS(12) - apo
Method: single particle / : Travis BA, Peck J, Schumacher MA

PDB-7tfe: 
L. monocytogenes GS(12) - apo
Method: single particle / : Travis BA, Peck J, Schumacher MA

EMDB-26105: 
BamABCDE bound to substrate EspP class 1
Method: single particle / : Doyle MT, Jimah JR, Dowdy T, Ohlemacher SI, Larion M, Hinshaw JE, Bernstein HD

EMDB-26106: 
BamABCDE bound to substrate EspP class 2
Method: single particle / : Doyle MT, Jimah JR, Dowdy T, Ohlemacher SI, Larion M, Hinshaw JE, Bernstein HD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
