-Search query
-Search result
Showing all 32 items for (author: kim & ys)
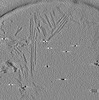
EMDB-28752: 
Cryo-electron tomography of wild type a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS
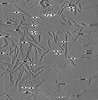
EMDB-28753: 
Cryo-electron tomography of S42Y a-synuclein preformed fibrils
Method: electron tomography / : Jiang J, Boparai N, Dai W, Kim YS

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
Method: subtomogram averaging / : Kaplan M, Ghosal D
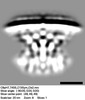
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D
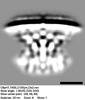
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D

EMDB-34530: 
Membrane protein A
Method: single particle / : Tajima S, Kim Y, Yamashita K, Fukuda M, Deisseroth K, Kato HE

EMDB-34531: 
Membrane protein B
Method: single particle / : Tajima S, Kim Y, Yamashita K, Fukuda M, Deisseroth K, Kato HE

EMDB-35713: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc
Method: single particle / : Tajima S, Kim Y, Nakamura S, Yamashita K, Fukuda M, Deisseroth K, Kato HE

EMDB-15270: 
SARS Cov2 Spike RBD in complex with Fab47
Method: single particle / : Hallberg BM, Das H

PDB-8a95: 
SARS Cov2 Spike RBD in complex with Fab47
Method: single particle / : Hallberg BM, Das H

EMDB-15273: 
SARS Cov2 Spike in 1-up conformation complex with Fab47
Method: single particle / : Hallberg BM, Das H

PDB-8a99: 
SARS Cov2 Spike in 1-up conformation complex with Fab47
Method: single particle / : Hallberg BM, Das H

EMDB-15269: 
SARS CoV2 Spike in the 2-up state in complex with Fab47
Method: single particle / : Hallberg BM, Das H

EMDB-15271: 
SARS Cov2 Spike RBD in complex with Fab47
Method: single particle / : Hallberg BM, Das H

PDB-8a94: 
SARS CoV2 Spike in the 2-up state in complex with Fab47.
Method: single particle / : Hallberg BM, Das H

PDB-8a96: 
SARS Cov2 Spike RBD in complex with Fab47
Method: single particle / : Hallberg BM, Das H

EMDB-26357: 
CryoEM structure of the Candida albicans Aro1 dimer
Method: single particle / : Quade B, Borek D, Otwinowski Z

EMDB-26358: 
Structure of DHQS/EPSPS dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z

EMDB-26359: 
Structure of the SK/DHQase/DHSD dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z

PDB-7u5s: 
CryoEM structure of the Candida albicans Aro1 dimer
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7u5t: 
Structure of DHQS/EPSPS dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-7u5u: 
Structure of the SK/DHQase/DHSD dimer from Candida albicans Aro1
Method: single particle / : Quade B, Borek D, Otwinowski Z, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-32377: 
2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine
Method: single particle / : Kishi KE, Kim Y, Fukuda M, Yamashita K, Deisseroth K, Kato HE

EMDB-32378: 
2.12 angstrom cryo-EM map of the pump-like channelrhodopsin ChRmine with Fab antibody fragment
Method: single particle / : Kishi KE, Kim Y, Fukuda M, Yamashita K, Deisseroth K, Kato HE

EMDB-23263: 
Cryo-EM map of pyridoxal 5'-phosphate synthase-like subunit PDX1.2 (Arabidopsis thaliana)
Method: single particle / : Novikova IV, Evans JE

EMDB-23264: 
Cryo-EM map of PDX1.2/PDX1.3 co-expression complex (Arabidopsis thaliana)
Method: single particle / : Novikova IV, Evans JE

PDB-7lb5: 
Pyridoxal 5'-phosphate synthase-like subunit PDX1.2 (Arabidopsis thaliana)
Method: single particle / : Novikova IV, Evans JE

PDB-7lb6: 
PDX1.2/PDX1.3 co-expression complex
Method: single particle / : Novikova IV, Evans JE

EMDB-20857: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

EMDB-20863: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

PDB-6ur8: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

PDB-6usf: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
