-Search query
-Search result
Showing 1 - 50 of 459 items for (author: ho & mr)

EMDB-41271: 
Consensus map of 96nm repeat of human respiratory doublet microtubule, RS1-2 region
Method: single particle / : Gui M, Brown A

EMDB-44496: 
Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor
Method: single particle / : Su CC

EMDB-44500: 
Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor
Method: single particle / : Su CC

EMDB-44501: 
Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor
Method: single particle / : Su CC
EMDB-44506: 
Cryo-EM co-structure of AcrB with CU244
Method: single particle / : Su CC

EMDB-40411: 
PHF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS
EMDB-40413: 
SF Tau from Down Syndrome
Method: helical / : Hoq MR, Bharath SR, Jiang W, Vago FS
EMDB-40416: 
Type I beta-amyloid 42 Filaments from Down syndrome
Method: helical / : Hoq MR, Bharath SR, Vago FS, Jiang W

EMDB-40419: 
Type IIIa beta-amyloid 40 Filaments from Down syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W
EMDB-40421: 
Type IIIb beta-amyloid 40 Filaments from Down Syndrome
Method: helical / : Hoq MR, Vago FS, Bharath SR, Jiang W

EMDB-19406: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-19407: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-41830: 
Lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41831: 
Gradient-fixed lipidated recombinant apolipoprotein E4
Method: single particle / : Strickland MR, Rau M, Summers B, Basore K, Wulf II J, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JAJ, Cashikar AG, Holtzman DM

EMDB-41138: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip
Method: single particle / : Kirk NS

EMDB-41837: 
The structure of the PP2A-B56Delta holoenzyme mutant - E197K
Method: single particle / : Wu CG, Xing Y

EMDB-42018: 
The structure of the PP2A-B56Delta holoenzyme mutant - E197K
Method: single particle / : Wu CG, Xing Y

EMDB-27141: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2)
Method: single particle / : Rai J, Das A, Li H
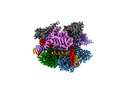
EMDB-27142: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1)
Method: single particle / : Rai J, Das A, Li H
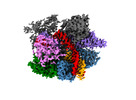
EMDB-27143: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage)
Method: single particle / : Rai J, Das A, Li H
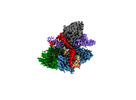
EMDB-27144: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2)
Method: single particle / : Rai J, Das A, Li H
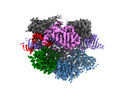
EMDB-27145: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound)
Method: single particle / : Rai J, Das A, Li H
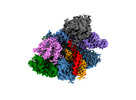
EMDB-27146: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1)
Method: single particle / : Rai J, Das A, Li H

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I

EMDB-16522: 
Structure of ADDoCoV-ADAH11
Method: single particle / : Yadav KNS, Buzas D, Berger-Schaffitzel C, Berger I

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle
Method: single particle / : Bufton JC, Capin J, Boruku U, Garzoni F, Schaffitzel C, Berger I

EMDB-41510: 
Eukaryotic translation initiation factor 2B tetramer
Method: single particle / : Wang L, Lawrence R, Sangwan S, Anand A, Shoemaker S, Deal A, Marqusee S, Watler P

EMDB-41566: 
Eukaryotic translation initiation factor 2B with a mutation (L516A) in the delta subunit
Method: single particle / : Wang L, Lawrence R, Sangwan S, Anand A, Shoemaker S, Deal A, Marqusee S, Watler P

PDB-8tqo: 
Eukaryotic translation initiation factor 2B tetramer
Method: single particle / : Wang L, Lawrence R, Sangwan S, Anand A, Shoemaker S, Deal A, Marqusee S, Watler P

PDB-8tqz: 
Eukaryotic translation initiation factor 2B with a mutation (L516A) in the delta subunit
Method: single particle / : Wang L, Lawrence R, Sangwan S, Anand A, Shoemaker S, Deal A, Marqusee S, Watler P
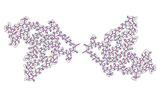
EMDB-28943: 
TMEM106B doublet filaments extracted from MSTD neurodegenerative human brain
Method: helical / : Hoq MR, Bharath SR, Jiang W

EMDB-41579: 
Structure of full-length LexA bound to a RecA filament
Method: helical / : Cory MB, Li A, Kohli RM

PDB-8trg: 
Structure of full-length LexA bound to a RecA filament
Method: helical / : Cory MB, Li A, Kohli RM

EMDB-36891: 
96nm repeat of human respiratory doublet microtubule, IDAf local refined
Method: single particle / : Gui M, Brown A

EMDB-36895: 
Consensus map of 96nm repeat of human respiratory doublet microtubule, RS3 region
Method: single particle / : Gui M, Brown A

EMDB-29980: 
Cryo-EM structure of serine 87 O-GlcNAc-modified alpha-synuclein fibrils
Method: helical / : Balana JA, Nguyen AB, Saelices L, Pratt RM

EMDB-41667: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body
Method: single particle / : Fuller JR, Padi SKR, Peti W, Page R

EMDB-41668: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body
Method: single particle / : Fuller JR, Padi SKR, Peti W, Page R

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
Method: single particle / : Duhoo Y, Lau K
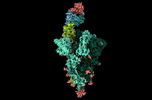
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)
Method: single particle / : Duhoo Y, Lau K

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.
Method: single particle / : Duhoo Y, Lau K

EMDB-40644: 
Cryo-EM structure of the PP2A:B55-FAM122A complex
Method: single particle / : Fuller JR, Padi SKR, Peti W, Page R

EMDB-41604: 
Cryo-EM structure of the PP2A:B55-ARPP19 complex
Method: single particle / : Fuller JR, Padi SKR, Peti W, Page R

EMDB-34000: 
Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34001: 
Spiral hexamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34002: 
Spiral pentamer of the substrate-free Lon protease with a S678A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34003: 
Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
