-Search query
-Search result
Showing 1 - 50 of 5,468 items for (author: david & l)

EMDB-42776: 
Triplet microtubule from the proximal region of basal body, WT, Tetrahymena thermophila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42777: 
Triplet microtubule from the basal body central core region, wildtype Tetrahymena thermophila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42778: 
Triplet microtubule from the distal region of basal body, wildtype Tetrahymena thermophila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42780: 
Triplet microtubule from the proximal region of basal body, focusing on the A/B inner junction, wildtype, Tetrahymena thermophila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42781: 
Triplet microtubule from the proximal region of basal body, focusing on the B/C inner junction, wildtype Tetrahymena themorphila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42782: 
Triplet microtubule from the central core region of basal body, focusing on the A/B inner junction, wildtype Tetrahymena thermophila
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42783: 
Triplet microtubule from the proximal region of basal body isolated from a Tetrahymena thermophila strain with POC1 knockout
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-42784: 
Triplet microtubule from the central core region of basal body isolated from a Tetrahymena thermophila mutant strain with POC1 knockout
Method: subtomogram averaging / : Ruehle M, Li S, Agard DA, Pearson C

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11
Method: single particle / : Karlinsey D, Ozorowski G, Ward AB

EMDB-29560: 
5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

EMDB-29571: 
4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

EMDB-29585: 
Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

EMDB-29597: 
LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

EMDB-29599: 
Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

PDB-8fy8: 
5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

PDB-8fye: 
4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

PDB-8fyl: 
Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

PDB-8fyt: 
LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D

PDB-8fyx: 
Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
Method: single particle / : Warren AL, Zilberg G, Capper MJ, Wacker D
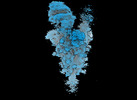
EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8qsq: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME

PDB-8qtd: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab
Method: single particle / : Ren J, Duyvesteyn HME, Stuart DI

PDB-8r8k: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-40940: 
TRPV1 in Nanodisc bound with lysophosphatidic acid in all four monomers
Method: single particle / : Arnold WR, Cheng Y

EMDB-40941: 
TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo)
Method: single particle / : Arnold WR, Cheng Y

EMDB-40949: 
TRPV1 in nanodisc bound with one LPA in one monomer
Method: single particle / : Arnold WR, Cheng Y

EMDB-40951: 
TRPV1 in nanodisc bound with two LPA molecules in opposite monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41005: 
TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41006: 
TRPV1 in nanodisc bound with 3 LPA molecules
Method: single particle / : Arnold WR, Julius D, Cheng Y
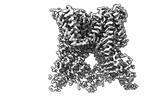
EMDB-41847: 
TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state
Method: single particle / : Arnold WR, Julius D, Cheng Y
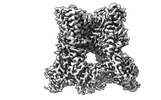
EMDB-41848: 
TRPV1 in nanodisc bound with diC8-PIP2 in the closed state
Method: single particle / : Arnold WR, Julius D, Cheng Y
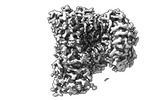
EMDB-41855: 
TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 1 (monomer)
Method: single particle / : Arnold WR, Julius D, Cheng Y
EMDB-41857: 
TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer)
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41864: 
TRPV1 in nanodisc bound with empty vanilloid binding pocket at 4C
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41866: 
TRPV1 in nanodisc bound with empty vanilloid binding pocket at 25C
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41873: 
TRPV1 in nanodisc bound with PIP2-Br4
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41879: 
TRPV1 in nanodisc bound with PI-Br4, consensus structure
Method: single particle / : Arnold WR, Julius D, Cheng Y

PDB-8t0c: 
TRPV1 in Nanodisc bound with lysophosphatidic acid in all four monomers
Method: single particle / : Arnold WR, Cheng Y

PDB-8t0e: 
TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo)
Method: single particle / : Arnold WR, Cheng Y

PDB-8t0y: 
TRPV1 in nanodisc bound with one LPA in one monomer
Method: single particle / : Arnold WR, Cheng Y

PDB-8t10: 
TRPV1 in nanodisc bound with two LPA molecules in opposite monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

PDB-8t3l: 
TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

PDB-8t3m: 
TRPV1 in nanodisc bound with 3 LPA molecules
Method: single particle / : Arnold WR, Julius D, Cheng Y

PDB-8u2z: 
TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state
Method: single particle / : Arnold WR, Julius D, Cheng Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
