+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9129 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of E.coli LptB2FGC with long BJR | |||||||||
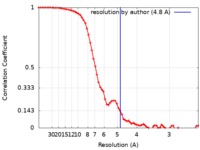
 Map data Map data | Cryo-EM map of LptB2FGC with long BJR, filtered to 4.8A with -150 b-factor | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Orlando BJ / Li Y / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structural basis of lipopolysaccharide extraction by the LptBFGC complex. Authors: Yanyan Li / Benjamin J Orlando / Maofu Liao /  Abstract: In Gram-negative bacteria, lipopolysaccharide is essential for outer membrane formation and antibiotic resistance. The seven lipopolysaccharide transport (Lpt) proteins A-G move lipopolysaccharide ...In Gram-negative bacteria, lipopolysaccharide is essential for outer membrane formation and antibiotic resistance. The seven lipopolysaccharide transport (Lpt) proteins A-G move lipopolysaccharide from the inner to the outer membrane. The ATP-binding cassette transporter LptBFG, which tightly associates with LptC, extracts lipopolysaccharide out of the inner membrane. The mechanism of the LptBFG-LptC complex (LptBFGC) and the role of LptC in lipopolysaccharide transport are poorly understood. Here we characterize the structures of LptBFG and LptBFGC in nucleotide-free and vanadate-trapped states, using single-particle cryo-electron microscopy. These structures resolve the bound lipopolysaccharide, reveal transporter-lipopolysaccharide interactions with side-chain details and uncover how the capture and extrusion of lipopolysaccharide are coupled to conformational rearrangements of LptBFGC. LptC inserts its transmembrane helix between the two transmembrane domains of LptBFG, which represents a previously unknown regulatory mechanism for ATP-binding cassette transporters. Our results suggest a role for LptC in achieving efficient lipopolysaccharide transport, by coordinating the action of LptBFG in the inner membrane and Lpt protein interactions in the periplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9129.map.gz emd_9129.map.gz | 24.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9129-v30.xml emd-9129-v30.xml emd-9129.xml emd-9129.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9129_fsc.xml emd_9129_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9129.png emd_9129.png | 56.1 KB | ||
| Others |  emd_9129_additional.map.gz emd_9129_additional.map.gz | 20.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9129 http://ftp.pdbj.org/pub/emdb/structures/EMD-9129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9129 | HTTPS FTP |
-Validation report
| Summary document |  emd_9129_validation.pdf.gz emd_9129_validation.pdf.gz | 78.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9129_full_validation.pdf.gz emd_9129_full_validation.pdf.gz | 77.7 KB | Display | |
| Data in XML |  emd_9129_validation.xml.gz emd_9129_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9129 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9129 | HTTPS FTP |
-Related structure data
| Related structure data |  9118C  9124C  9125C  9126C  9128C  9130C  6mhuC  6mhzC  6mi7C  6mi8C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9129.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9129.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of LptB2FGC with long BJR, filtered to 4.8A with -150 b-factor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unfiltered cryo-EM map of LptB2FGC with long BJR
| File | emd_9129_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered cryo-EM map of LptB2FGC with long BJR | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LptB2FGC
| Entire | Name: LptB2FGC |
|---|---|
| Components |
|
-Supramolecule #1: LptB2FGC
| Supramolecule | Name: LptB2FGC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
| Details | nanodisc incorporated LptB2FGC |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)