+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9125 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleotide-free Cryo-EM Structure of E.coli LptB2FGC | |||||||||
 Map data Map data | Cryo-EM map of nucleotide-free LptB2FGC filtered to 4.2A with -200 b-factor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / lipopolysaccharide / LPS / nanodisc / MEMBRANE PROTEIN / HYDROLASE-TRANSPORT PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationlipopolysaccharide transmembrane transporter activity / Translocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate / glycolipid transfer activity / transporter complex / lipopolysaccharide transport / Gram-negative-bacterium-type cell outer membrane assembly / ATP-binding cassette (ABC) transporter complex / transmembrane transport / outer membrane-bounded periplasmic space / ATP hydrolysis activity ...lipopolysaccharide transmembrane transporter activity / Translocases; Catalysing the translocation of carbohydrates and their derivatives; Linked to the hydrolysis of a nucleoside triphosphate / glycolipid transfer activity / transporter complex / lipopolysaccharide transport / Gram-negative-bacterium-type cell outer membrane assembly / ATP-binding cassette (ABC) transporter complex / transmembrane transport / outer membrane-bounded periplasmic space / ATP hydrolysis activity / ATP binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
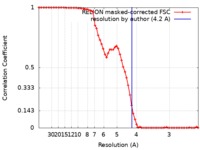
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Orlando BJ / Li Y | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structural basis of lipopolysaccharide extraction by the LptBFGC complex. Authors: Yanyan Li / Benjamin J Orlando / Maofu Liao /  Abstract: In Gram-negative bacteria, lipopolysaccharide is essential for outer membrane formation and antibiotic resistance. The seven lipopolysaccharide transport (Lpt) proteins A-G move lipopolysaccharide ...In Gram-negative bacteria, lipopolysaccharide is essential for outer membrane formation and antibiotic resistance. The seven lipopolysaccharide transport (Lpt) proteins A-G move lipopolysaccharide from the inner to the outer membrane. The ATP-binding cassette transporter LptBFG, which tightly associates with LptC, extracts lipopolysaccharide out of the inner membrane. The mechanism of the LptBFG-LptC complex (LptBFGC) and the role of LptC in lipopolysaccharide transport are poorly understood. Here we characterize the structures of LptBFG and LptBFGC in nucleotide-free and vanadate-trapped states, using single-particle cryo-electron microscopy. These structures resolve the bound lipopolysaccharide, reveal transporter-lipopolysaccharide interactions with side-chain details and uncover how the capture and extrusion of lipopolysaccharide are coupled to conformational rearrangements of LptBFGC. LptC inserts its transmembrane helix between the two transmembrane domains of LptBFG, which represents a previously unknown regulatory mechanism for ATP-binding cassette transporters. Our results suggest a role for LptC in achieving efficient lipopolysaccharide transport, by coordinating the action of LptBFG in the inner membrane and Lpt protein interactions in the periplasm. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9125.map.gz emd_9125.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9125-v30.xml emd-9125-v30.xml emd-9125.xml emd-9125.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9125_fsc.xml emd_9125_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_9125.png emd_9125.png | 77.7 KB | ||
| Filedesc metadata |  emd-9125.cif.gz emd-9125.cif.gz | 7.3 KB | ||
| Others |  emd_9125_additional.map.gz emd_9125_additional.map.gz | 20.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9125 http://ftp.pdbj.org/pub/emdb/structures/EMD-9125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9125 | HTTPS FTP |
-Related structure data
| Related structure data |  6mi7MC  9118C  9124C  9126C  9128C  9129C  9130C  6mhuC  6mhzC  6mi8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9125.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9125.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of nucleotide-free LptB2FGC filtered to 4.2A with -200 b-factor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
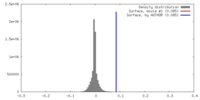
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unfiltered map of nucleotide-free LptB2FGC
| File | emd_9125_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered map of nucleotide-free LptB2FGC | ||||||||||||
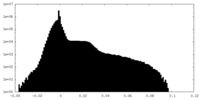
| Projections & Slices |
| ||||||||||||
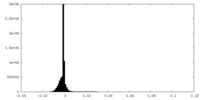
| Density Histograms |
- Sample components
Sample components
-Entire : LptB2FGC
| Entire | Name: LptB2FGC |
|---|---|
| Components |
|
-Supramolecule #1: LptB2FGC
| Supramolecule | Name: LptB2FGC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lipopolysaccharide export system permease protein LptF
| Macromolecule | Name: Lipopolysaccharide export system permease protein LptF type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.393473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPANLVLS LLGLGVPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLV KAAMILAVFT AIVAAVNVMW AGPWSSRHQD EVLAEAKANP GMAALAQGQF QQATNGSSVL F IESVDGSD ...String: MIIIRYLVRE TLKSQLAILF ILLLIFFCQK LVRILGAAVD GDIPANLVLS LLGLGVPEMA QLILPLSLFL GLLMTLGKLY TESEITVMH ACGLSKAVLV KAAMILAVFT AIVAAVNVMW AGPWSSRHQD EVLAEAKANP GMAALAQGQF QQATNGSSVL F IESVDGSD FKDVFLAQIR PKGNARPSVV VADSGHLTQL RDGSQVVTLN QGTRFEGTAL LRDFRITDFQ DYQAIIGHQA VA LDPNDTD QMDMRTLWNT DTDRARAELN WRITLVFTVF MMALMVVPLS VVNPRQGRVL SMLPAMLLYL LFFLIQTSLK SNG GKGKLD PTLWMWTVNL IYLALAIVLN LWDTVPVRRL RASFSRKGAV UniProtKB: Lipopolysaccharide export system permease protein LptF |
-Macromolecule #2: Lipopolysaccharide export system permease protein LptG
| Macromolecule | Name: Lipopolysaccharide export system permease protein LptG type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.65141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQPFGVLDRY IGKTIFTTIM MTLFMLVSLS GIIKFVDQLK KAGQGSYDAL GAGMYTLLSV PKDVQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR MQVALSVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQA MYGGSLLSTQ QGLWAKDGNN F VYIERVKG ...String: MQPFGVLDRY IGKTIFTTIM MTLFMLVSLS GIIKFVDQLK KAGQGSYDAL GAGMYTLLSV PKDVQIFFPM AALLGALLGL GMLAQRSEL VVMQASGFTR MQVALSVMKT AIPLVLLTMA IGEWVAPQGE QMARNYRAQA MYGGSLLSTQ QGLWAKDGNN F VYIERVKG DEELGGISIY AFNENRRLQS VRYAATAKFD PEHKVWRLSQ VDESDLTNPK QITGSQTVSG TWKTNLTPDK LG VVALDPD ALSISGLHNY VKYLKSSGQD AGRYQLNMWS KIFQPLSVAV MMLMALSFIF GPLRSVPMGV RVVTGISFGF VFY VLDQIF GPLTLVYGIP PIIGALLPSA SFFLISLWLL MRKS UniProtKB: Lipopolysaccharide export system permease protein LptG |
-Macromolecule #3: Lipopolysaccharide export system ATP-binding protein LptB
| Macromolecule | Name: Lipopolysaccharide export system ATP-binding protein LptB type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.131088 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDDDISLL PLHARARRG IGYLPQEASI FRRLSVYDNL MAVLQIRDDL SAEQREDRAN ELMEEFHIEH LRDSMGQSLS GGERRRVEIA R ALAANPKF ...String: MGHHHHHHHH MATLTAKNLA KAYKGRRVVE DVSLTVNSGE IVGLLGPNGA GKTTTFYMVV GIVPRDAGNI IIDDDDISLL PLHARARRG IGYLPQEASI FRRLSVYDNL MAVLQIRDDL SAEQREDRAN ELMEEFHIEH LRDSMGQSLS GGERRRVEIA R ALAANPKF ILLDEPFAGV DPISVIDIKR IIEHLRDSGL GVLITDHNVR ETLAVCERAY IVSQGHLIAH GTPTEILQDE HV KRVYLGE DFRL UniProtKB: Lipopolysaccharide export system ATP-binding protein LptB |
-Macromolecule #4: Lipopolysaccharide export system protein LptC
| Macromolecule | Name: Lipopolysaccharide export system protein LptC / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.808877 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKE NLYFQGGMSK ARRWVIIVLS LAVLVMIGIN MAEKDDTAQV VVNNNDPTYK SEHTDTLVYN PEGALSYRLI AQHVEYYSD QAVSWFTQPV LTTFDKDKIP TWSVKADKAK LTNDRMLYLY GHVEVNALVP DSQLRRITTD NAQINLVTQD V TSEDLVTL ...String: MWSHPQFEKE NLYFQGGMSK ARRWVIIVLS LAVLVMIGIN MAEKDDTAQV VVNNNDPTYK SEHTDTLVYN PEGALSYRLI AQHVEYYSD QAVSWFTQPV LTTFDKDKIP TWSVKADKAK LTNDRMLYLY GHVEVNALVP DSQLRRITTD NAQINLVTQD V TSEDLVTL YGTTFNSSGL KMRGNLRSKN AELIEKVRTS YEIQNKQTQP UniProtKB: Lipopolysaccharide export system protein LptC |
-Macromolecule #5: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 5 / Number of copies: 1 / Formula: PGT |
|---|---|
| Molecular weight | Theoretical: 751.023 Da |
| Chemical component information |  ChemComp-PGT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
| Details | nanodisc incorporated LptB2FGC |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)