+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21307 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NatB complex | |||||||||
 Map data Map data | NatB complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NatB / NAA20 / NAA25 / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationN-terminal peptidyl-glutamine acetylation / N-terminal methionine Nalpha-acetyltransferase NatB / N-terminal peptidyl-aspartic acid acetylation / N-terminal peptidyl-glutamic acid acetylation / NatB complex / N-terminal protein amino acid acetylation / protein N-terminal-methionine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / acetyltransferase activator activity / cytoskeleton organization ...N-terminal peptidyl-glutamine acetylation / N-terminal methionine Nalpha-acetyltransferase NatB / N-terminal peptidyl-aspartic acid acetylation / N-terminal peptidyl-glutamic acid acetylation / NatB complex / N-terminal protein amino acid acetylation / protein N-terminal-methionine acetyltransferase activity / protein-N-terminal amino-acid acetyltransferase activity / acetyltransferase activator activity / cytoskeleton organization / regulation of actin cytoskeleton organization / Golgi apparatus / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Deng S / Marmorstein R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Molecular basis for N-terminal alpha-synuclein acetylation by human NatB. Authors: Sunbin Deng / Buyan Pan / Leah Gottlieb / E James Petersson / Ronen Marmorstein /  Abstract: NatB is one of three major N-terminal acetyltransferase (NAT) complexes (NatA-NatC), which co-translationally acetylate the N-termini of eukaryotic proteins. Its substrates account for about 21% of ...NatB is one of three major N-terminal acetyltransferase (NAT) complexes (NatA-NatC), which co-translationally acetylate the N-termini of eukaryotic proteins. Its substrates account for about 21% of the human proteome, including well known proteins such as actin, tropomyosin, CDK2, and α-synuclein (αSyn). Human NatB (hNatB) mediated N-terminal acetylation of αSyn has been demonstrated to play key roles in the pathogenesis of Parkinson's disease and as a potential therapeutic target for hepatocellular carcinoma. Here we report the cryo-EM structure of hNatB bound to a CoA-αSyn conjugate, together with structure-guided analysis of mutational effects on catalysis. This analysis reveals functionally important differences with human NatA and NatB, resolves key hNatB protein determinants for αSyn N-terminal acetylation, and identifies important residues for substrate-specific recognition and acetylation by NatB enzymes. These studies have implications for developing small molecule NatB probes and for understanding the mode of substrate selection by NAT enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21307.map.gz emd_21307.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21307-v30.xml emd-21307-v30.xml emd-21307.xml emd-21307.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21307.png emd_21307.png | 106 KB | ||
| Filedesc metadata |  emd-21307.cif.gz emd-21307.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21307 http://ftp.pdbj.org/pub/emdb/structures/EMD-21307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21307 | HTTPS FTP |
-Related structure data
| Related structure data |  6vp9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10477 (Title: Cryo-EM structure of Human NatB with an Alpha-Synuclein peptide and CoA conjugate EMPIAR-10477 (Title: Cryo-EM structure of Human NatB with an Alpha-Synuclein peptide and CoA conjugateData size: 2.0 TB Data #1: unaligned raw movies of hNatB with an Alpha-Synuclein peptide and CoA conjugate [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21307.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21307.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NatB complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human NatB complex
| Entire | Name: human NatB complex |
|---|---|
| Components |
|
-Supramolecule #1: human NatB complex
| Supramolecule | Name: human NatB complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: N-alpha-acetyltransferase 20, N-alpha-acetyltransferase 25, NatB ...
| Supramolecule | Name: N-alpha-acetyltransferase 20, N-alpha-acetyltransferase 25, NatB auxiliary subunit type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: MDVFM peptide
| Supramolecule | Name: MDVFM peptide / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: N-alpha-acetyltransferase 20
| Macromolecule | Name: N-alpha-acetyltransferase 20 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: N-terminal methionine Nalpha-acetyltransferase NatB |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.694168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTLRAFTCD DLFRFNNINL DPLTETYGIP FYLQYLAHWP EYFIVAEAPG GELMGYIMGK AEGSVAREEW HGHVTALSVA PEFRRLGLA AKLMELLEEI SERKGGFFVD LFVRVSNQVA VNMYKQLGYS VYRTVIEYYS ASNGEPDEDA YDMRKALSRD T EKK UniProtKB: N-alpha-acetyltransferase 20 |
-Macromolecule #2: N-alpha-acetyltransferase 25, NatB auxiliary subunit
| Macromolecule | Name: N-alpha-acetyltransferase 25, NatB auxiliary subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 112.444258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATRGHVQDP NDRRLRPIYD YLDNGNNKMA IQQADKLLKK HKDLHCAKVL KAIGLQRTGK QEEAFTLAQE VAALEPTDDN SLQALTILY REMHRPELVT KLYEAAVKKV PNSEEYHSHL FMAYARVGEY KKMQQAGMAL YKIVPKNPYY FWSVMSLIMQ S ISAQDENL ...String: MATRGHVQDP NDRRLRPIYD YLDNGNNKMA IQQADKLLKK HKDLHCAKVL KAIGLQRTGK QEEAFTLAQE VAALEPTDDN SLQALTILY REMHRPELVT KLYEAAVKKV PNSEEYHSHL FMAYARVGEY KKMQQAGMAL YKIVPKNPYY FWSVMSLIMQ S ISAQDENL SKTMFLPLAE RMVEKMVKED KIEAEAEVEL YYMILERLGK YQEALDVIRG KLGEKLTSEI QSRENKCMAM YK KLSRWPE CNALSRRLLL KNSDDWQFYL TYFDSVFRLI EEAWSPPAEG EHSLEGEVHY SAEKAVKFIE DRITEESKSS RHL RGPHLA KLELIRRLRS QGCNDEYKLG DPEELMFQYF KKFGDKPCCF TDLKVFVDLL PATQCTKFIN QLLGVVPLST PTED KLALP ADIRALQQHL CVVQLTRLLG LYHTMDKNQK LSVVRELMLR YQHGLEFGKT CLKTELQFSD YYCLLAVHAL IDVWR ETGD ETTVWQALTL LEEGLTHSPS NAQFKLLLVR IYCMLGAFEP VVDLYSSLDA KHIQHDTIGY LLTRYAESLG QYAAAS QSC NFALRFFHSN QKDTSEYIIQ AYKYGAFEKI PEFIAFRNRL NNSLHFAQVR TERMLLDLLL EANISTSLAE SIKSMNL RP EEDDIPWEDL RDNRDLNVFF SWDPKDRDVS EEHKKLSLEE ETLWLRIRSL TLRLISGLPS LNHPVEPKNS EKTAENGV S SRIDILRLLL QQLEATLETG KRFIEKDIQY PFLGPVPTRM GGFFNSGCSQ CQISSFYLVN DIYELDTSGL EDTMEIQER IENSFKSLLD QLKDVFSKCK GDLLEVKDGN LKTHPTLLEN LVFFVETISV ILWVSSYCES VLRPYKLNLQ KKKKKKKETS IIMPPVFTS FQDYVTGLQT LISNVVDHIK GLETHLIALK LEELILEDTS LSPEERKFSK TVQGKVQSSY LHSLLEMGEL L KKRLETTK KLKI UniProtKB: N-alpha-acetyltransferase 25, NatB auxiliary subunit |
-Macromolecule #3: MDVFM peptide
| Macromolecule | Name: MDVFM peptide / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 641.799 Da |
| Sequence | String: MDVFM |
-Macromolecule #4: CARBOXYMETHYL COENZYME *A
| Macromolecule | Name: CARBOXYMETHYL COENZYME *A / type: ligand / ID: 4 / Number of copies: 1 / Formula: CMC |
|---|---|
| Molecular weight | Theoretical: 825.57 Da |
| Chemical component information | 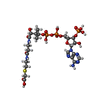 ChemComp-CMC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Average electron dose: 1.6 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6vp9: |
 Movie
Movie Controller
Controller



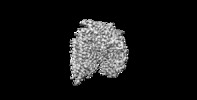















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















