[English] 日本語
 Yorodumi
Yorodumi- EMDB-7941: Fako virus empty particles aligned to the best decoy map (asymmet... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7941 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fako virus empty particles aligned to the best decoy map (asymmetric reconstruction) showing the locations of the polymerase complexes | |||||||||
 Map data Map data | Asymmetric reconstruction of Fako virus empty particles aligned to the best decoy map, showing the locations of the polymerase complexes | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Fako virus Fako virus | |||||||||
| Method | single particle reconstruction / cryo EM | |||||||||
 Authors Authors | Kaelber JT / Jiang W / Weaver SC / Auguste AJ / Chiu W | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Arrangement of the Polymerase Complexes inside a Nine-Segmented dsRNA Virus. Authors: Jason T Kaelber / Wen Jiang / Scott C Weaver / Albert J Auguste / Wah Chiu /  Abstract: Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle ...Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle reconstruction encounters difficulties resolving structures such as the intraparticle polymerase complex because refinement can converge to an incorrect map and because the map could depict a nonrepresentative subset of particles or an average of heterogeneous particles. Using the nine-segmented Fako virus, we tested hypotheses for the arrangement and number of polymerase complexes within the virion by measuring how well each hypothesis describes the set of cryoelectron microscopy images of individual viral particles. We find that the polymerase complex in Fako virus binds at ten possible sites despite having only nine genome segments. A single asymmetric configuration describes the arrangement of these complexes in both virions and genome-free capsids. Similarities between the arrangements of Reoviridae with 9, 10, and 11 segments indicate the generalizability of this architecture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7941.map.gz emd_7941.map.gz | 240.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7941-v30.xml emd-7941-v30.xml emd-7941.xml emd-7941.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7941.png emd_7941.png | 118.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7941 http://ftp.pdbj.org/pub/emdb/structures/EMD-7941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7941 | HTTPS FTP |
-Validation report
| Summary document |  emd_7941_validation.pdf.gz emd_7941_validation.pdf.gz | 77.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7941_full_validation.pdf.gz emd_7941_full_validation.pdf.gz | 77.1 KB | Display | |
| Data in XML |  emd_7941_validation.xml.gz emd_7941_validation.xml.gz | 499 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7941 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7941 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7941 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7941 | HTTPS FTP |
-Related structure data
| Related structure data |  7944C  7945C  7948C  7949C  7953C  7954C  6djyC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7941.map.gz / Format: CCP4 / Size: 253.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7941.map.gz / Format: CCP4 / Size: 253.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric reconstruction of Fako virus empty particles aligned to the best decoy map, showing the locations of the polymerase complexes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.142 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
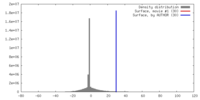
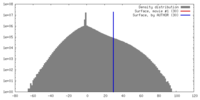
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fako virus
| Entire | Name:   Fako virus Fako virus |
|---|---|
| Components |
|
-Supramolecule #1: Fako virus
| Supramolecule | Name: Fako virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1567404 / Sci species name: Fako virus / Sci species strain: CSW77 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Culicinae (mosquito) Culicinae (mosquito) |
| Molecular weight | Theoretical: 43 MDa |
| Virus shell | Shell ID: 1 / T number (triangulation number): 2 |
-Macromolecule #1: major capsid protein
| Macromolecule | Name: major capsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKI ETNRSSQDSV QHGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS T SHGREPEI ESFADRAELA MMIQGMTVGA LTVQPMRSIR STFANLANVL ...String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKI ETNRSSQDSV QHGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS T SHGREPEI ESFADRAELA MMIQGMTVGA LTVQPMRSIR STFANLANVL IFHDVFTTED KP SAFIEYH SDEMIVNMPK QTYNPIDNLA KILYLPSLEK FKYGTGIVQL NYSPHISKLY QNT NNIINT ITDGITYANR TEFFIRVMVL MMMDRKILTM EFYDVDTSAI SNTAILPTIP TTTG VSPLL RIDTRTEPIW YNDAIKTLIT NLTIQYGKIK TVLDANAVKR YSVVGYPIDQ YRAYL YNHN LLEYLGKKVK REDIMSLIKA LSYEFDLITI SDLEYQNIPK WFSDNDLSRF IFSICM FPD IVRQFHALNI DYFSQANVFT VKSENAIVKM LNSNQNMEPT IINWFLFRIC AIDKTVI DD YFSLEMTPII MRPKLYDFDM KRGEPVSLLY ILELILFSIM FPNVTQHMLG QIQARILY I SMYAFRQEYL KFITKFGFYY KIVNGRKEYI QVTNQNERMT ENNDVLTGNL YPSLFTDDP TLSAIAPTLA KIARLMKPTT SLTPDDRAIA AKFPRFKDSA HLNPYSSLNI GGRTQHSVTY TRMYDAIEE MFNLILRAFA SSFAQRPRAG VTQLKSLLTQ LADPLCLALD GHVYHLYNVM A NMMQNFIP NTDGQFHSFR ACSYAVKDGG NIYRVVQNGD ELNESLLIDT AIVWGLLGNT DS SYGNAIG ATGTANVPTK VQPVIPTPDN FITPTIHLKT SIDAICSVEG ILLLILSRQT TIP GYEDEL NKLRTGISQP KVTERQYRRA RESIKNMLGS GDYNVAPLHF LLHTEHRSTK LSKP LIRRV LDNVVQPYVA NLDPAEFENT PQLIENSNMT RLQIALKMLT GDMDDIVKGL ILHKR ACAK FDVYETLTIP TDVKTIVLTM QHISTQTQNN MVYYVFLIDG VKILAEDIKN VNFQID ITG IWPEYVITLL LRAINNGFNT YVSMPNILYK PTITADVRQF MNTTKAETLL ISNKSIV HE IMFFDNALQP KMSSDTLALS EAVYRTIWNS SIITQRISAR GLMNLEDARP PEAKISHQ S ELDMGKIDET SGEPIYTSGL QKMQSSKVSM ANVVLSAGSD VIRQAAIKYN VVRTQEIIL FE |
-Macromolecule #2: RNA-dependent RNA polymerase
| Macromolecule | Name: RNA-dependent RNA polymerase / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MATEPNEINL RMTQTLMKQI EDTTFKIRRK HDDMYTYLID DTAAIEMYPD TETPFRMKSS LHELYIQVK CQIPKSIIRP PLYNNDMYPL NNNNTNFLED RPFNYLCNFD WYEFLSRTKD E LGMHYNMI RDLIAMSSQT RYSNIWSNIS GIIMYLRQYQ RGFALRAILA ...String: MATEPNEINL RMTQTLMKQI EDTTFKIRRK HDDMYTYLID DTAAIEMYPD TETPFRMKSS LHELYIQVK CQIPKSIIRP PLYNNDMYPL NNNNTNFLED RPFNYLCNFD WYEFLSRTKD E LGMHYNMI RDLIAMSSQT RYSNIWSNIS GIIMYLRQYQ RGFALRAILA RLLKQWNNFP FF DTWDGFR DVLPNTSTAW PLLFYAFMSV TFDYICENIS EGEAIVCFQN HLENAQVSYQ DTK LEKKNN YTLWLKYECH NLRVALTPRY DYTGTVSGNF LKDTTEYIIE NEFENNEIAM RNNL LPSFY AILQQMRQKL KTVEDIIRLL TICYSARDDR TYYGTLMELA ISKAIKPQVA GSIVP APIP TSWLQKDPKM VLSARYPSTS FLSQMQEFYR RYYPALQKEI DVHALSASFI NFLSTA SAG VGIELPEEIV NMVQDKRLLY LIKKGSGKRV LQEALLVEKY NDLDPIINDF VRLIKIV VR KQIERRQRGI AGIPNNVLKI NQVTYEANKP FSKIARAPSH GKQSGNASDI HDLLFYTT Q EDVSHIETNG KRQMRGIVIS SADVKGMDTH IQINAAMNQH LGAIEILDGI QYDVGPFRQ TNAIIQDIQG NVYEKSLNGG QQAIAFGLAN FSQTTGINSK YFGQIPNQEG TFPSGLITTS NHHTQMLTL LIETALTTFT KEFGKSMAIS HLMILGDDVS LMLHGNDKDI NFFMKYLVEK F SQLGLILE RDESRNFGVF LQQHVINGRF NGFSNRIAIF TSEDYKTRKS VRESCTEYNA LI DDVIFRT YNVRKLLQFQ RIHQFVVLSK YVFRIQNYKY ESLRAKLAKR MNVFEYELKI KDN DKTDVH NRNALRFIGI QIPYTYFQYS GGGEIPPESF QKKDGSFTYE YSIYSPRGKW LRKF LYDIS HDARDVKFQI DHEMMKLYNL DICDFLLQYN VLDIQEEIRS TIIDRELVSK LAMNL ESLE HSNARMISRR ASESLRSMGI RLPANGVYGY QINERLVKVL QNIQQSDYEV KMVGDT LFT AIMEKFEHHK VRMEKGDKLH NFTLDFSNKD DKITINRKML ALHNISISKN MMPYSDA WM LYTCLDNTYN TSSDLATALA HSQGHFKSFQ YDRDMFAESV KIASKHGIGS LPMELFFE A SNIRDTAQLK WIEAIKYYVQ FKDYLYPYSL NPRRLFFIPE QVSSVSNILN QENIPADIN QKALLFRRAY AYVLSHPFCI SGAKAIFVED RIS |
-Macromolecule #3: turret
| Macromolecule | Name: turret / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAI SEIDDALDEF IQTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN N PQLLNYLD DNPFSAIFEL VNVDLQIYQY GQNIFNNEAE HTILFLKDNT ...String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAI SEIDDALDEF IQTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN N PQLLNYLD DNPFSAIFEL VNVDLQIYQY GQNIFNNEAE HTILFLKDNT NYGVIQALQK HP FSATHIN WHLHKHIFVF HSREQLLNKL LSAGLEDSQL YQRQKTYSTK RGDRPTERMV TYI EDDHIR RIQAVFPLLL DNIFDVKLHK DSSMTWLKSY ADMIYDSVKN SNSTITPEIR KLYL RMYNQ YMRIFLPIEQ YMLYDNTCWP FSEKITLKIN VRLISSRENQ PVLWKTPIDT ENLIS IVQP DEPINKLNFT AIPSTMIRLN DNITMYRAVK DMFSAIEYLP DAIENIPTLT MKEQAL SRY ISPDSEAQNF FNNQPPYLNS IMNVNRQVFE AVKRGNIQVS TGSMEHLCLC MHVKSGL IV GRTVLIDDKV VLRRNFNAST AKMITCYVKA FAQLYGEGSL INPGLRMVFF GVETEPAI D ILKLFYGDKS LYIQGFGDRG IGRDKFRTKI EDALTLRIGC DILISDIDQA DYEDPNEEK FDDITDFVCY VTELVISNAT VGLVKISMPT YYIMNKISST LNNKFSNVAI NIVKLSTQKP YTYEAYIML SHGSTLTNKG YLRNPVCDVY LEKISLQPMD LKIISTISNE INYDKPTLYR F VVDKNDVT DVSIAMHILS IHCSTITTRS VMVRSDNTGA FVTMSGIKDM KRVAIMNRMT DG TSANSYM HEQNGKLYLQ KVPYLEDLIS AFPNGFGSTY QNDYDSSMSV INVNALIRQV VYR VISKSI PVALLESLSR IRIIGGRDLG EMNAVYKLYK TPIEVYDAVG ITREYPHVQI SYRA QRYSF TESIPNHTLL LANYVIMNDV DGAPISSLEQ INTIKKIISK ISLGSIAYIQ VYTDI VARN INVMTKNDSF LISANADKTV FKVQVSGYKA VEMCNYEQLL QLVSDNTGVN IIKLTY QDV LESCVLSSGI LGDTGSWLLD LVLASTYIIE IRG |
-Macromolecule #4: clamp
| Macromolecule | Name: clamp / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDF DRDNQLAMAH FPDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS I NLVRQPPM DNVMYWACQF LSSGTSFLPL ERDVEIVFSG FKGSHICMFS ...String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDF DRDNQLAMAH FPDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS I NLVRQPPM DNVMYWACQF LSSGTSFLPL ERDVEIVFSG FKGSHICMFS NLRQMNLSPI LC PYYDLIT NFKTTTEIRA YVDAHEELKS LLTYLCLCTI VGLCDTFTET RNMDTGEYVW KVR DVVSRN HTPAQNVEKF CYTIQNAKYM IQLVHVLLFP LTDNKYADLP NYVAVITQGA INQS RSHNV INTTDESNSN TTSDTAASTS GIVSGDTGTV ASLYPDEFKY VQS |
-Macromolecule #5: accessory NTPase
| Macromolecule | Name: accessory NTPase / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MFAIPFTASQ VYSHLGLYQQ PIEKLDPLTK VQYESAEKLR DIITQLSRRS GGISLRKNLI RDIAFGNKP KLNIPSLTHQ VRHNREFKHG RGQFILVDSN GSDYYDGIFD DMVLILRSNF V LPNVPGII SEQLRLIVID NDFYLKHIPS EIAILRDVLS EHMVKVSIDG ...String: MFAIPFTASQ VYSHLGLYQQ PIEKLDPLTK VQYESAEKLR DIITQLSRRS GGISLRKNLI RDIAFGNKP KLNIPSLTHQ VRHNREFKHG RGQFILVDSN GSDYYDGIFD DMVLILRSNF V LPNVPGII SEQLRLIVID NDFYLKHIPS EIAILRDVLS EHMVKVSIDG VSLDDPYISK FF RKPFILY DHTKFRKVDL TKVVLINKTG KSKAQLIDKY KLTDDAVFIS YEVFDIMLQP VKS RDGDIE QFVRMRGDIE TWKMKIGTTF ESNLMELFIH GVPVMNSTSQ LQLDFQISDI RSAN VFDEQ IVCAAKLNGL REQCLKESSL IRVNIIGQKG SGKSMLMRLI NERGIPGISR KIICI DSDA YGKWKTQTQY LDDKFSALKL INVDNLHEIS QDDNIISYYE QFIIDQLLAH NITISE HTN SIRFIKQFKV DTLKDIGRKF KELYLADFKE QIHFYEYLCG NIPEPSSTLL ITFLHAT VE TSAAPGTNMN FSLNTILYPL QSILNRKRGA LVNFILNRIY DEMGTEAFTR LRPCDVWK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Pretreatment - Type: PLASMA CLEANING / Details: unspecified | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Number real images: 2400 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.7 mm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)