[English] 日本語
 Yorodumi
Yorodumi- EMDB-7954: Fako virus full particles, projection-matching reconstruction wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7954 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fako virus full particles, projection-matching reconstruction without imposed symmetry | |||||||||
 Map data Map data | Unsymmetrized reconstruction of Fako virus full particles | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Fako virus Fako virus | |||||||||
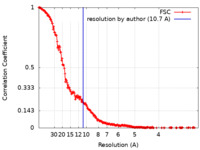
| Method | single particle reconstruction / cryo EM / Resolution: 10.7 Å | |||||||||
 Authors Authors | Kaelber JT / Jiang W / Weaver SC / Auguste AJ / Chiu W | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Arrangement of the Polymerase Complexes inside a Nine-Segmented dsRNA Virus. Authors: Jason T Kaelber / Wen Jiang / Scott C Weaver / Albert J Auguste / Wah Chiu /  Abstract: Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle ...Members of the family Reoviridae package several copies of the viral polymerase complex into their capsid to carry out replication and transcription within viral particles. Classical single-particle reconstruction encounters difficulties resolving structures such as the intraparticle polymerase complex because refinement can converge to an incorrect map and because the map could depict a nonrepresentative subset of particles or an average of heterogeneous particles. Using the nine-segmented Fako virus, we tested hypotheses for the arrangement and number of polymerase complexes within the virion by measuring how well each hypothesis describes the set of cryoelectron microscopy images of individual viral particles. We find that the polymerase complex in Fako virus binds at ten possible sites despite having only nine genome segments. A single asymmetric configuration describes the arrangement of these complexes in both virions and genome-free capsids. Similarities between the arrangements of Reoviridae with 9, 10, and 11 segments indicate the generalizability of this architecture. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7954.map.gz emd_7954.map.gz | 328.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7954-v30.xml emd-7954-v30.xml emd-7954.xml emd-7954.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7954_fsc.xml emd_7954_fsc.xml | 20.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_7954.png emd_7954.png | 233 KB | ||
| Others |  emd_7954_half_map_1.map.gz emd_7954_half_map_1.map.gz emd_7954_half_map_2.map.gz emd_7954_half_map_2.map.gz | 328.2 MB 328.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7954 http://ftp.pdbj.org/pub/emdb/structures/EMD-7954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7954 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7954 | HTTPS FTP |
-Related structure data
| Related structure data |  7941C  7944C  7945C  7948C  7949C  7953C  6djyC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7954.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7954.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsymmetrized reconstruction of Fako virus full particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
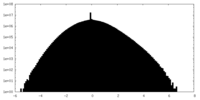
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.506 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
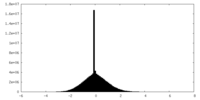
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Unsymmetrized reconstruction of Fako virus full particles. Unmasked,...
| File | emd_7954_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsymmetrized reconstruction of Fako virus full particles. Unmasked, unfiltered half-set map. | ||||||||||||
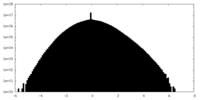
| Projections & Slices |
| ||||||||||||
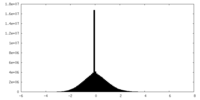
| Density Histograms |
-Half map: Unsymmetrized reconstruction of Fako virus full particles. Unmasked,...
| File | emd_7954_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsymmetrized reconstruction of Fako virus full particles. Unmasked, unfiltered half-set map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fako virus
| Entire | Name:   Fako virus Fako virus |
|---|---|
| Components |
|
-Supramolecule #1: Fako virus
| Supramolecule | Name: Fako virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1567404 / Sci species name: Fako virus / Sci species strain: CSW77 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Culicinae (mosquito) Culicinae (mosquito) |
| Molecular weight | Theoretical: 43 MDa |
| Virus shell | Shell ID: 1 / T number (triangulation number): 2 |
-Macromolecule #1: clamp protein
| Macromolecule | Name: clamp protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDFD RDNQLAMAH FPDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS INLVRQPPMD NVMYWACQFL S SGTSFLPL ERDVEIVFSG FKGSHICMFS ...String: MTLTYWDKEK RMTLKQMIQQ VAINEQENEL THYVFTTPLS MPTFGKPMLG YVPLNEVATS KFFSNVNDFD RDNQLAMAH FPDTTITQAY NLTNSIKPGD TSLPDAEVAA LKWFWKFFTS INLVRQPPMD NVMYWACQFL S SGTSFLPL ERDVEIVFSG FKGSHICMFS NLRQMNLSPI LCPYYDLITN FKTTTEIRAY VDAHEELKSL LT YLCLCTI VGLCDTFTET RNMDTGEYVW KVRDVVSRNH TPAQNVEKFC YTIQNAKYMI QLVHVLLFPL TDN KYADLP NYVAVITQGA INQSRSHNVI NTTDESNSNT TSDTAASTSG IVSGDTGTVA SLYPDEFKYV QS |
-Macromolecule #2: major capsid protein
| Macromolecule | Name: major capsid protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKIE TNRSSQDSV QHGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS TSHGREPEIE SFADRAELAM M IQGMTVGA LTVQPMRSIR STFANLANVL ...String: MRPIRMYKNN QERTNLKHQE INEEQQNEQT TSNQGFTRSD NSGKINIERI SSSRNQITDG KTVSSYSKIE TNRSSQDSV QHGGSSITYT SDTTGNPRIT NARTNNDETH ATGPIEDLNS TSHGREPEIE SFADRAELAM M IQGMTVGA LTVQPMRSIR STFANLANVL IFHDVFTTED KPSAFIEYHS DEMIVNMPKQ TYNPIDNLAK IL YLPSLEK FKYGTGIVQL NYSPHISKLY QNTNNIINTI TDGITYANRT EFFIRVMVLM MMDRKILTME FYD VDTSAI SNTAILPTIP TTTGVSPLLR IDTRTEPIWY NDAIKTLITN LTIQYGKIKT VLDANAVKRY SVVG YPIDQ YRAYLYNHNL LEYLGKKVKR EDIMSLIKAL SYEFDLITIS DLEYQNIPKW FSDNDLSRFI FSICM FPDI VRQFHALNID YFSQANVFTV KSENAIVKML NSNQNMEPTI INWFLFRICA IDKTVIDDYF SLEMTP IIM RPKLYDFDMK RGEPVSLLYI LELILFSIMF PNVTQHMLGQ IQARILYISM YAFRQEYLKF ITKFGFY YK IVNGRKEYIQ VTNQNERMTE NNDVLTGNLY PSLFTDDPTL SAIAPTLAKI ARLMKPTTSL TPDDRAIA A KFPRFKDSAH LNPYSSLNIG GRTQHSVTYT RMYDAIEEMF NLILRAFASS FAQRPRAGVT QLKSLLTQL ADPLCLALDG HVYHLYNVMA NMMQNFIPNT DGQFHSFRAC SYAVKDGGNI YRVVQNGDEL NESLLIDTAI VWGLLGNTD SSYGNAIGAT GTANVPTKVQ PVIPTPDNFI TPTIHLKTSI DAICSVEGIL LLILSRQTTI P GYEDELNK LRTGISQPKV TERQYRRARE SIKNMLGSGD YNVAPLHFLL HTEHRSTKLS KPLIRRVLDN VV QPYVANL DPAEFENTPQ LIENSNMTRL QIALKMLTGD MDDIVKGLIL HKRACAKFDV YETLTIPTDV KTI VLTMQH ISTQTQNNMV YYVFLIDGVK ILAEDIKNVN FQIDITGIWP EYVITLLLRA INNGFNTYVS MPNI LYKPT ITADVRQFMN TTKAETLLIS NKSIVHEIMF FDNALQPKMS SDTLALSEAV YRTIWNSSII TQRIS ARGL MNLEDARPPE AKISHQSELD MGKIDETSGE PIYTSGLQKM QSSKVSMANV VLSAGSDVIR QAAIKY NVV RTQEIILFE |
-Macromolecule #3: turret protein
| Macromolecule | Name: turret protein / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAIS EIDDALDEF IQTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN NPQLLNYLDD NPFSAIFELV N VDLQIYQY GQNIFNNEAE HTILFLKDNT ...String: MIDLRLEEDI LTATLPEFLS TRPKYRYAYT NTKQQDIRFQ GPMRHVRLTH LYKQTKLWNL QYIERELAIS EIDDALDEF IQTFSLPYVI EQGTYKYNML LGMHAHNVNY QDDVSELIAN NPQLLNYLDD NPFSAIFELV N VDLQIYQY GQNIFNNEAE HTILFLKDNT NYGVIQALQK HPFSATHINW HLHKHIFVFH SREQLLNKLL SA GLEDSQL YQRQKTYSTK RGDRPTERMV TYIEDDHIRR IQAVFPLLLD NIFDVKLHKD SSMTWLKSYA DMI YDSVKN SNSTITPEIR KLYLRMYNQY MRIFLPIEQY MLYDNTCWPF SEKITLKINV RLISSRENQP VLWK TPIDT ENLISIVQPD EPINKLNFTA IPSTMIRLND NITMYRAVKD MFSAIEYLPD AIENIPTLTM KEQAL SRYI SPDSEAQNFF NNQPPYLNSI MNVNRQVFEA VKRGNIQVST GSMEHLCLCM HVKSGLIVGR TVLIDD KVV LRRNFNASTA KMITCYVKAF AQLYGEGSLI NPGLRMVFFG VETEPAIDIL KLFYGDKSLY IQGFGDR GI GRDKFRTKIE DALTLRIGCD ILISDIDQAD YEDPNEEKFD DITDFVCYVT ELVISNATVG LVKISMPT Y YIMNKISSTL NNKFSNVAIN IVKLSTQKPY TYEAYIMLSH GSTLTNKGYL RNPVCDVYLE KISLQPMDL KIISTISNEI NYDKPTLYRF VVDKNDVTDV SIAMHILSIH CSTITTRSVM VRSDNTGAFV TMSGIKDMKR VAIMNRMTD GTSANSYMHE QNGKLYLQKV PYLEDLISAF PNGFGSTYQN DYDSSMSVIN VNALIRQVVY R VISKSIPV ALLESLSRIR IIGGRDLGEM NAVYKLYKTP IEVYDAVGIT REYPHVQISY RAQRYSFTES IP NHTLLLA NYVIMNDVDG APISSLEQIN TIKKIISKIS LGSIAYIQVY TDIVARNINV MTKNDSFLIS ANA DKTVFK VQVSGYKAVE MCNYEQLLQL VSDNTGVNII KLTYQDVLES CVLSSGILGD TGSWLLDLVL ASTY IIEIR G |
-Macromolecule #4: RNA-dependent RNA polymerase
| Macromolecule | Name: RNA-dependent RNA polymerase / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MATEPNEINL RMTQTLMKQI EDTTFKIRRK HDDMYTYLID DTAAIEMYPD TETPFRMKSS LHELYIQVKC QIPKSIIRP PLYNNDMYPL NNNNTNFLED RPFNYLCNFD WYEFLSRTKD ELGMHYNMIR DLIAMSSQTR Y SNIWSNIS GIIMYLRQYQ RGFALRAILA ...String: MATEPNEINL RMTQTLMKQI EDTTFKIRRK HDDMYTYLID DTAAIEMYPD TETPFRMKSS LHELYIQVKC QIPKSIIRP PLYNNDMYPL NNNNTNFLED RPFNYLCNFD WYEFLSRTKD ELGMHYNMIR DLIAMSSQTR Y SNIWSNIS GIIMYLRQYQ RGFALRAILA RLLKQWNNFP FFDTWDGFRD VLPNTSTAWP LLFYAFMSVT FD YICENIS EGEAIVCFQN HLENAQVSYQ DTKLEKKNNY TLWLKYECHN LRVALTPRYD YTGTVSGNFL KDT TEYIIE NEFENNEIAM RNNLLPSFYA ILQQMRQKLK TVEDIIRLLT ICYSARDDRT YYGTLMELAI SKAI KPQVA GSIVPAPIPT SWLQKDPKMV LSARYPSTSF LSQMQEFYRR YYPALQKEID VHALSASFIN FLSTA SAGV GIELPEEIVN MVQDKRLLYL IKKGSGKRVL QEALLVEKYN DLDPIINDFV RLIKIVVRKQ IERRQR GIA GIPNNVLKIN QVTYEANKPF SKIARAPSHG KQSGNASDIH DLLFYTTQED VSHIETNGKR QMRGIVI SS ADVKGMDTHI QINAAMNQHL GAIEILDGIQ YDVGPFRQTN AIIQDIQGNV YEKSLNGGQQ AIAFGLAN F SQTTGINSKY FGQIPNQEGT FPSGLITTSN HHTQMLTLLI ETALTTFTKE FGKSMAISHL MILGDDVSL MLHGNDKDIN FFMKYLVEKF SQLGLILERD ESRNFGVFLQ QHVINGRFNG FSNRIAIFTS EDYKTRKSVR ESCTEYNAL IDDVIFRTYN VRKLLQFQRI HQFVVLSKYV FRIQNYKYES LRAKLAKRMN VFEYELKIKD N DKTDVHNR NALRFIGIQI PYTYFQYSGG GEIPPESFQK KDGSFTYEYS IYSPRGKWLR KFLYDISHDA RD VKFQIDH EMMKLYNLDI CDFLLQYNVL DIQEEIRSTI IDRELVSKLA MNLESLEHSN ARMISRRASE SLR SMGIRL PANGVYGYQI NERLVKVLQN IQQSDYEVKM VGDTLFTAIM EKFEHHKVRM EKGDKLHNFT LDFS NKDDK ITINRKMLAL HNISISKNMM PYSDAWMLYT CLDNTYNTSS DLATALAHSQ GHFKSFQYDR DMFAE SVKI ASKHGIGSLP MELFFEASNI RDTAQLKWIE AIKYYVQFKD YLYPYSLNPR RLFFIPEQVS SVSNIL NQE NIPADINQKA LLFRRAYAYV LSHPFCISGA KAIFVEDRI |
-Macromolecule #5: polymerase accessory protein
| Macromolecule | Name: polymerase accessory protein / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Fako virus / Strain: CSW77 Fako virus / Strain: CSW77 |
| Sequence | String: MFAIPFTASQ VYSHLGLYQQ PIEKLDPLTK VQYESAEKLR DIITQLSRRS GGISLRKNLI RDIAFGNKPK LNIPSLTHQ VRHNREFKHG RGQFILVDSN GSDYYDGIFD DMVLILRSNF VLPNVPGIIS EQLRLIVIDN D FYLKHIPS EIAILRDVLS EHMVKVSIDG ...String: MFAIPFTASQ VYSHLGLYQQ PIEKLDPLTK VQYESAEKLR DIITQLSRRS GGISLRKNLI RDIAFGNKPK LNIPSLTHQ VRHNREFKHG RGQFILVDSN GSDYYDGIFD DMVLILRSNF VLPNVPGIIS EQLRLIVIDN D FYLKHIPS EIAILRDVLS EHMVKVSIDG VSLDDPYISK FFRKPFILYD HTKFRKVDLT KVVLINKTGK SK AQLIDKY KLTDDAVFIS YEVFDIMLQP VKSRDGDIEQ FVRMRGDIET WKMKIGTTFE SNLMELFIHG VPV MNSTSQ LQLDFQISDI RSANVFDEQI VCAAKLNGLR EQCLKESSLI RVNIIGQKGS GKSMLMRLIN ERGI PGISR KIICIDSDAY GKWKTQTQYL DDKFSALKLI NVDNLHEISQ DDNIISYYEQ FIIDQLLAHN ITISE HTNS IRFIKQFKVD TLKDIGRKFK ELYLADFKEQ IHFYEYLCGN IPEPSSTLLI TFLHATVETS AAPGTN MNF SLNTILYPLQ SILNRKRGAL VNFILNRIYD EMGTEAFTRL RPCDVWK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Number real images: 2400 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.7 mm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)