[English] 日本語
 Yorodumi
Yorodumi- EMDB-7579: SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7579 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | ||||||||||||
 Map data Map data | primary map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | membrane fusion / glycoprotein / receptor binding / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / endocytosis involved in viral entry into host cell / SARS-CoV-1 activates/modulates innate immune responses / suppression by virus of host tetherin activity / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / positive regulation of viral entry into host cell ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / Attachment and Entry / endocytosis involved in viral entry into host cell / SARS-CoV-1 activates/modulates innate immune responses / suppression by virus of host tetherin activity / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  SARS coronavirus / SARS coronavirus /  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | ||||||||||||
 Authors Authors | Kirchdoerfer RN / Wang N | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Authors: Robert N Kirchdoerfer / Nianshuang Wang / Jesper Pallesen / Daniel Wrapp / Hannah L Turner / Christopher A Cottrell / Kizzmekia S Corbett / Barney S Graham / Jason S McLellan / Andrew B Ward /  Abstract: Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 as a highly transmissible pathogenic human betacoronavirus. The viral spike glycoprotein (S) utilizes angiotensin-converting ...Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 as a highly transmissible pathogenic human betacoronavirus. The viral spike glycoprotein (S) utilizes angiotensin-converting enzyme 2 (ACE2) as a host protein receptor and mediates fusion of the viral and host membranes, making S essential to viral entry into host cells and host species tropism. As SARS-CoV enters host cells, the viral S is believed to undergo a number of conformational transitions as it is cleaved by host proteases and binds to host receptors. We recently developed stabilizing mutations for coronavirus spikes that prevent the transition from the pre-fusion to post-fusion states. Here, we present cryo-EM analyses of a stabilized trimeric SARS-CoV S, as well as the trypsin-cleaved, stabilized S, and its interactions with ACE2. Neither binding to ACE2 nor cleavage by trypsin at the S1/S2 cleavage site impart large conformational changes within stabilized SARS-CoV S or expose the secondary cleavage site, S2'. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7579.map.gz emd_7579.map.gz | 116.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7579-v30.xml emd-7579-v30.xml emd-7579.xml emd-7579.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7579_fsc.xml emd_7579_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_7579.png emd_7579.png | 155.9 KB | ||
| Masks |  emd_7579_msk_1.map emd_7579_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-7579.cif.gz emd-7579.cif.gz | 7.3 KB | ||
| Others |  emd_7579_half_map_1.map.gz emd_7579_half_map_1.map.gz emd_7579_half_map_2.map.gz emd_7579_half_map_2.map.gz | 98.4 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7579 http://ftp.pdbj.org/pub/emdb/structures/EMD-7579 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7579 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7579 | HTTPS FTP |
-Validation report
| Summary document |  emd_7579_validation.pdf.gz emd_7579_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7579_full_validation.pdf.gz emd_7579_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_7579_validation.xml.gz emd_7579_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_7579_validation.cif.gz emd_7579_validation.cif.gz | 24 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7579 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7579 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7579 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7579 | HTTPS FTP |
-Related structure data
| Related structure data |  6cs1MC  7573C  7574C  7575C  7576C  7577C  7578C  7580C  7581C  7582C  7584C  7585C  7586C  7601C  7602C  7603C  7604C  7605C  7606C  7607C  7608C  6crvC  6crwC  6crxC  6crzC  6cs0C  6cs2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7579.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7579.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7579_msk_1.map emd_7579_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
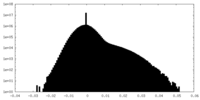
| Density Histograms |
-Half map: em-half-volume P1
| File | emd_7579_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
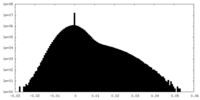
| Density Histograms |
-Half map: em-half-volume P2
| File | emd_7579_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS Spike Glycoprotein,Trypsin-cleaved, stabilized variant
| Entire | Name: SARS Spike Glycoprotein,Trypsin-cleaved, stabilized variant |
|---|---|
| Components |
|
-Supramolecule #1: SARS Spike Glycoprotein,Trypsin-cleaved, stabilized variant
| Supramolecule | Name: SARS Spike Glycoprotein,Trypsin-cleaved, stabilized variant type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  SARS coronavirus / Strain: Tor2 SARS coronavirus / Strain: Tor2 |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 134.739266 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SDLDRCTTFD DVQAPNYTQH TSSMRGVYYP DEIFRSDTLY LTQDLFLPFY SNVTGFHTIN HTFGNPVIPF KDGIYFAATE KSNVVRGWV FGSTMNNKSQ SVIIINNSTN VVIRACNFEL CDNPFFAVSK PMGTQTHTMI FDNAFNCTFE YISDAFSLDV S EKSGNFKH ...String: SDLDRCTTFD DVQAPNYTQH TSSMRGVYYP DEIFRSDTLY LTQDLFLPFY SNVTGFHTIN HTFGNPVIPF KDGIYFAATE KSNVVRGWV FGSTMNNKSQ SVIIINNSTN VVIRACNFEL CDNPFFAVSK PMGTQTHTMI FDNAFNCTFE YISDAFSLDV S EKSGNFKH LREFVFKNKD GFLYVYKGYQ PIDVVRDLPS GFNTLKPIFK LPLGINITNF RAILTAFSPA QDIWGTSAAA YF VGYLKPT TFMLKYDENG TITDAVDCSQ NPLAELKCSV KSFEIDKGIY QTSNFRVVPS GDVVRFPNIT NLCPFGEVFN ATK FPSVYA WERKKISNCV ADYSVLYNST FFSTFKCYGV SATKLNDLCF SNVYADSFVV KGDDVRQIAP GQTGVIADYN YKLP DDFMG CVLAWNTRNI DATSTGNYNY KYRYLRHGKL RPFERDISNV PFSPDGKPCT PPALNCYWPL NDYGFYTTTG IGYQP YRVV VLSFELLNAP ATVCGPKLST DLIKNQCVNF NFNGLTGTGV LTPSSKRFQP FQQFGRDVSD FTDSVRDPKT SEILDI SPC AFGGVSVITP GTNASSEVAV LYQDVNCTDV STAIHADQLT PAWRIYSTGN NVFQTQAGCL IGAEHVDTSY ECDIPIG AG ICASYHTVSL LRSTSQKSIV AYTMSLGADS SIAYSNNTIA IPTNFSISIT TEVMPVSMAK TSVDCNMYIC GDSTECAN L LLQYGSFCTQ LNRALSGIAA EQDRNTREVF AQVKQMYKTP TLKYFGGFNF SQILPDPLKP TKRSFIEDLL FNKVTLADA GFMKQYGECL GDINARDLIC AQKFNGLTVL PPLLTDDMIA AYTAALVSGT ATAGWTFGAG AALQIPFAMQ MAYRFNGIGV TQNVLYENQ KQIANQFNKA ISQIQESLTT TSTALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE V QIDRLITG RLQSLQTYVT QQLIRAAEIR ASANLAATKM SECVLGQSKR VDFCGKGYHL MSFPQAAPHG VVFLHVTYVP SQ ERNFTTA PAICHEGKAY FPREGVFVFN GTSWFITQRN FFSPQIITTD NTFVSGNCDV VIGIINNTVY DPLQPELDSF KEE LDKYFK NHTSPDVDLG DISGINASVV NIQKEIDRLN EVAKNLNESL IDLQELGKYE QGSGYIPEAP RDGQAYVRKD GEWV LLSTF LGRSLEVLFQ UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: C-flat-2/2 4C / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 0.25 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-6cs1: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)