[English] 日本語
 Yorodumi
Yorodumi- EMDB-20829: MERS-CoV S structure in complex with 5-N-glycolyl neuraminic acid -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20829 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MERS-CoV S structure in complex with 5-N-glycolyl neuraminic acid | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Park YJ / Walls AC / Wang Z / Sauer MM / Li W / Tortorici MA / Bosch BJ / DiMaio F / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Authors: Young-Jun Park / Alexandra C Walls / Zhaoqian Wang / Maximillian M Sauer / Wentao Li / M Alejandra Tortorici / Berend-Jan Bosch / Frank DiMaio / David Veesler /    Abstract: The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe and often lethal respiratory illness in humans, and no vaccines or specific treatments are available. Infections are ...The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe and often lethal respiratory illness in humans, and no vaccines or specific treatments are available. Infections are initiated via binding of the MERS-CoV spike (S) glycoprotein to sialosides and dipeptidyl-peptidase 4 (the attachment and entry receptors, respectively). To understand MERS-CoV engagement of sialylated receptors, we determined the cryo-EM structures of S in complex with 5-N-acetyl neuraminic acid, 5-N-glycolyl neuraminic acid, sialyl-Lewis, α2,3-sialyl-N-acetyl-lactosamine and α2,6-sialyl-N-acetyl-lactosamine at 2.7-3.0 Å resolution. We show that recognition occurs via a conserved groove that is essential for MERS-CoV S-mediated attachment to sialosides and entry into human airway epithelial cells. Our data illuminate MERS-CoV S sialoside specificity and suggest that selectivity for α2,3-linked over α2,6-linked receptors results from enhanced interactions with the former class of oligosaccharides. This study provides a structural framework explaining MERS-CoV attachment to sialoside receptors and identifies a site of potential vulnerability to inhibitors of viral entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20829.map.gz emd_20829.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20829-v30.xml emd-20829-v30.xml emd-20829.xml emd-20829.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20829.png emd_20829.png | 54 KB | ||
| Others |  emd_20829_additional.map.gz emd_20829_additional.map.gz emd_20829_additional_1.map.gz emd_20829_additional_1.map.gz | 123 MB 123 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20829 http://ftp.pdbj.org/pub/emdb/structures/EMD-20829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20829 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20829.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20829.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_20829_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
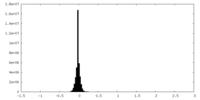
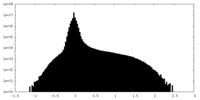
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_20829_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
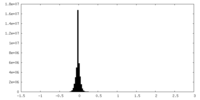
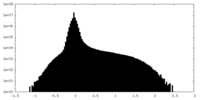
| Density Histograms |
- Sample components
Sample components
-Entire : MERS-CoV S ectodomain in complex with 5-N-glycolyl neuraminic acid
| Entire | Name: MERS-CoV S ectodomain in complex with 5-N-glycolyl neuraminic acid |
|---|---|
| Components |
|
-Supramolecule #1: MERS-CoV S ectodomain in complex with 5-N-glycolyl neuraminic acid
| Supramolecule | Name: MERS-CoV S ectodomain in complex with 5-N-glycolyl neuraminic acid type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 23281 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)