[English] 日本語
 Yorodumi
Yorodumi- EMDB-20544: MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20544 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coronavirus / spike glycoprotein / MERS-CoV / membrane fusion / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationreceptor-mediated endocytosis of virus by host cell / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane ...receptor-mediated endocytosis of virus by host cell / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |   Human betacoronavirus 2c EMC/2012 Human betacoronavirus 2c EMC/2012 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Park YJ / Walls AC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Authors: Young-Jun Park / Alexandra C Walls / Zhaoqian Wang / Maximillian M Sauer / Wentao Li / M Alejandra Tortorici / Berend-Jan Bosch / Frank DiMaio / David Veesler /    Abstract: The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe and often lethal respiratory illness in humans, and no vaccines or specific treatments are available. Infections are ...The Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe and often lethal respiratory illness in humans, and no vaccines or specific treatments are available. Infections are initiated via binding of the MERS-CoV spike (S) glycoprotein to sialosides and dipeptidyl-peptidase 4 (the attachment and entry receptors, respectively). To understand MERS-CoV engagement of sialylated receptors, we determined the cryo-EM structures of S in complex with 5-N-acetyl neuraminic acid, 5-N-glycolyl neuraminic acid, sialyl-Lewis, α2,3-sialyl-N-acetyl-lactosamine and α2,6-sialyl-N-acetyl-lactosamine at 2.7-3.0 Å resolution. We show that recognition occurs via a conserved groove that is essential for MERS-CoV S-mediated attachment to sialosides and entry into human airway epithelial cells. Our data illuminate MERS-CoV S sialoside specificity and suggest that selectivity for α2,3-linked over α2,6-linked receptors results from enhanced interactions with the former class of oligosaccharides. This study provides a structural framework explaining MERS-CoV attachment to sialoside receptors and identifies a site of potential vulnerability to inhibitors of viral entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20544.map.gz emd_20544.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20544-v30.xml emd-20544-v30.xml emd-20544.xml emd-20544.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20544.png emd_20544.png | 53.8 KB | ||
| Filedesc metadata |  emd-20544.cif.gz emd-20544.cif.gz | 6.8 KB | ||
| Others |  emd_20544_additional.map.gz emd_20544_additional.map.gz | 123.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20544 http://ftp.pdbj.org/pub/emdb/structures/EMD-20544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20544 | HTTPS FTP |
-Related structure data
| Related structure data |  6q06MC  6q04C  6q05C  6q07C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20544.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20544.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
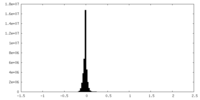
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_20544_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
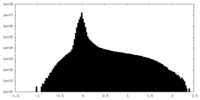
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MERS-CoV S ectodomain in complex with 5-N-acetyl neuraminic acid
| Entire | Name: MERS-CoV S ectodomain in complex with 5-N-acetyl neuraminic acid |
|---|---|
| Components |
|
-Supramolecule #1: MERS-CoV S ectodomain in complex with 5-N-acetyl neuraminic acid
| Supramolecule | Name: MERS-CoV S ectodomain in complex with 5-N-acetyl neuraminic acid type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human betacoronavirus 2c EMC/2012 Human betacoronavirus 2c EMC/2012 |
| Molecular weight | Theoretical: 149.172062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTVDVGPDSV KSACIEVDIQ QTFFDKTWPR PIDVSKADGI IYPQGRTYSN ITITYQGLF PYQGDHGDMY VYSAGHATGT TPQKLFVANY SQDVKQFANG FVVRIGAAAN STGTVIISPS TSATIRKIYP A FMLGSSVG ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTVDVGPDSV KSACIEVDIQ QTFFDKTWPR PIDVSKADGI IYPQGRTYSN ITITYQGLF PYQGDHGDMY VYSAGHATGT TPQKLFVANY SQDVKQFANG FVVRIGAAAN STGTVIISPS TSATIRKIYP A FMLGSSVG NFSDGKMGRF FNHTLVLLPD GCGTLLRAFY CILEPRSGNH CPAGNSYTSF ATYHTPATDC SDGNYNRNAS LN SFKEYFN LRNCTFMYTY NITEDEILEW FGITQTAQGV HLFSSRYVDL YGGNMFQFAT LPVYDTIKYY SIIPHSIRSI QSD RKAWAA FYVYKLQPLT FLLDFSVDGY IRRAIDCGFN DLSQLHCSYE SFDVESGVYS VSSFEAKPSG SVVEQAEGVE CDFS PLLSG TPPQVYNFKR LVFTNCNYNL TKLLSLFSVN DFTCSQISPA AIASNCYSSL ILDYFSYPLS MKSDLSVSSA GPISQ FNYK QSFSNPTCLI LATVPHNLTT ITKPLKYSYI NKCSRLLSDD RTEVPQLVNA NQYSPCVSIV PSTVWEDGDY YRKQLS PLE GGGWLVASGS TVAMTEQLQM GFGITVQYGT DTNSVCPKLE FANDTKIASQ LGNCVEYSLY GVSGRGVFQN CTAVGVR QQ RFVYDAYQNL VGYYSDDGNY YCLRACVSVP VSVIYDKETK THATLFGSVA CEHISSTMSQ YSRSTRSMLK RRDSTYGP L QTPVGCVLGL VNSSLFVEDC KLPLGQSLCA LPDTPSTLTP ASVGSVPGEM RLASIAFNHP IQVDQLNSSY FKLSIPTNF SFGVTQEYIQ TTIQKVTVDC KQYVCNGFQK CEQLLREYGQ FCSKINQALH GANLRQDDSV RNLFASVKSS QSSPIIPGFG GDFNLTLLE PVSISTGSRS ARSAIEDLLF DKVTIADPGY MQGYDDCMQQ GPASARDLIC AQYVAGYKVL PPLMDVNMEA A YTSSLLGS IAGVGWTAGL SSFAAIPFAQ SIFYRLNGVG ITQQVLSENQ KLIANKFNQA LGAMQTGFTT TNEAFQKVQD AV NNNAQAL SKLASELSNT FGAISASIGD IIQRLDPPEQ DAQIDRLING RLTTLNAFVA QQLVRSESAA LSAQLAKDKV NEC VKAQSK RSGFCGQGTH IVSFVVNAPN GLYFMHVGYY PSNHIEVVSA YGLCDAANPT NCIAPVNGYF IKTNNTRIVD EWSY TGSSF YAPEPITSLN TKYVAPQVTY QNISTNLPPP LLGNSTGIDF QDELDEFFKN VSTSIPNFGS LTQINTTLLD LTYEM LSLQ QVVKALNESY IDLKELGNYT YYNKGSGREN LYFQGGGGSG YIPEAPRDGQ AYVRKDGEWV LLSTFLGHHH HHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #9: FOLIC ACID
| Macromolecule | Name: FOLIC ACID / type: ligand / ID: 9 / Number of copies: 3 / Formula: FOL |
|---|---|
| Molecular weight | Theoretical: 441.397 Da |
| Chemical component information |  ChemComp-FA: |
-Macromolecule #10: water
| Macromolecule | Name: water / type: ligand / ID: 10 / Number of copies: 216 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 34458 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)