+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rat GluN1a-2B Fab 003-102 local refinement | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Channel / heterotetramer / receptor / antibody / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / pons maturation / positive regulation of Schwann cell migration / regulation of cell communication / regulation of cAMP/PKA signal transduction / EPHB-mediated forward signaling / sensitization ...cellular response to curcumin / cellular response to corticosterone stimulus / cellular response to magnesium starvation / sensory organ development / pons maturation / positive regulation of Schwann cell migration / regulation of cell communication / regulation of cAMP/PKA signal transduction / EPHB-mediated forward signaling / sensitization / auditory behavior / Assembly and cell surface presentation of NMDA receptors / response to hydrogen sulfide / olfactory learning / conditioned taste aversion / dendritic branch / regulation of respiratory gaseous exchange / response to other organism / protein localization to postsynaptic membrane / regulation of ARF protein signal transduction / transmitter-gated monoatomic ion channel activity / fear response / apical dendrite / positive regulation of inhibitory postsynaptic potential / response to methylmercury / response to carbohydrate / response to glycine / propylene metabolic process / response to manganese ion / interleukin-1 receptor binding / cellular response to dsRNA / response to glycoside / positive regulation of glutamate secretion / negative regulation of dendritic spine maintenance / cellular response to lipid / response to growth hormone / regulation of monoatomic cation transmembrane transport / heterocyclic compound binding / NMDA glutamate receptor activity / Synaptic adhesion-like molecules / RAF/MAP kinase cascade / voltage-gated monoatomic cation channel activity / neurotransmitter receptor complex / NMDA selective glutamate receptor complex / ligand-gated sodium channel activity / glutamate binding / response to morphine / regulation of axonogenesis / calcium ion transmembrane import into cytosol / neuromuscular process / regulation of dendrite morphogenesis / regulation of synapse assembly / protein heterotetramerization / male mating behavior / glycine binding / response to amine / parallel fiber to Purkinje cell synapse / receptor clustering / small molecule binding / suckling behavior / regulation of long-term synaptic depression / startle response / positive regulation of reactive oxygen species biosynthetic process / monoatomic cation transmembrane transport / behavioral response to pain / positive regulation of calcium ion transport into cytosol / response to magnesium ion / associative learning / regulation of postsynaptic membrane potential / regulation of MAPK cascade / action potential / cellular response to glycine / extracellularly glutamate-gated ion channel activity / monoatomic cation transport / excitatory synapse / social behavior / positive regulation of dendritic spine maintenance / positive regulation of excitatory postsynaptic potential / response to electrical stimulus / monoatomic ion channel complex / regulation of neuronal synaptic plasticity / cellular response to manganese ion / long-term memory / glutamate receptor binding / Unblocking of NMDA receptors, glutamate binding and activation / response to mechanical stimulus / behavioral fear response / detection of mechanical stimulus involved in sensory perception of pain / multicellular organismal response to stress / synaptic cleft / neuron development / prepulse inhibition / postsynaptic density, intracellular component / phosphatase binding / monoatomic cation channel activity / calcium ion homeostasis / response to fungicide / glutamate-gated receptor activity / D2 dopamine receptor binding / regulation of neuron apoptotic process Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Michalski K / Furukawa H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis. Authors: Kevin Michalski / Taha Abdulla / Sam Kleeman / Lars Schmidl / Ricardo Gómez / Noriko Simorowski / Francesca Vallese / Harald Prüss / Manfred Heckmann / Christian Geis / Hiro Furukawa /   Abstract: Autoantibodies against neuronal membrane proteins can manifest in autoimmune encephalitis, inducing seizures, cognitive dysfunction and psychosis. Anti-N-methyl-D-aspartate receptor (NMDAR) ...Autoantibodies against neuronal membrane proteins can manifest in autoimmune encephalitis, inducing seizures, cognitive dysfunction and psychosis. Anti-N-methyl-D-aspartate receptor (NMDAR) encephalitis is the most dominant autoimmune encephalitis; however, insights into how autoantibodies recognize and alter receptor functions remain limited. Here we determined structures of human and rat NMDARs bound to three distinct patient-derived antibodies using single-particle electron cryo-microscopy. These antibodies bind different regions within the amino-terminal domain of the GluN1 subunit. Through electrophysiology, we show that all three autoantibodies acutely and directly reduced NMDAR channel functions in primary neurons. Antibodies show different stoichiometry of binding and antibody-receptor complex formation, which in one antibody, 003-102, also results in reduced synaptic localization of NMDARs. These studies demonstrate mechanisms of diverse epitope recognition and direct channel regulation of anti-NMDAR autoantibodies underlying autoimmune encephalitis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43559.map.gz emd_43559.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43559-v30.xml emd-43559-v30.xml emd-43559.xml emd-43559.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43559.png emd_43559.png | 69.4 KB | ||
| Filedesc metadata |  emd-43559.cif.gz emd-43559.cif.gz | 6.6 KB | ||
| Others |  emd_43559_half_map_1.map.gz emd_43559_half_map_1.map.gz emd_43559_half_map_2.map.gz emd_43559_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43559 http://ftp.pdbj.org/pub/emdb/structures/EMD-43559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43559 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43559 | HTTPS FTP |
-Validation report
| Summary document |  emd_43559_validation.pdf.gz emd_43559_validation.pdf.gz | 937.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43559_full_validation.pdf.gz emd_43559_full_validation.pdf.gz | 937.5 KB | Display | |
| Data in XML |  emd_43559_validation.xml.gz emd_43559_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_43559_validation.cif.gz emd_43559_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43559 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43559 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43559 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43559 | HTTPS FTP |
-Related structure data
| Related structure data |  8vvhMC  8vuhC  8vujC  8vulC  8vunC  8vuqC  8vurC  8vusC  8vutC  8vuuC  8vuvC  8vuyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43559.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43559.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
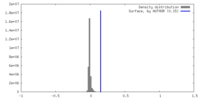
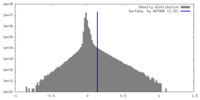
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43559_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
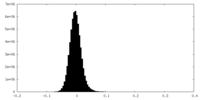
| Density Histograms |
-Half map: #2
| File | emd_43559_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
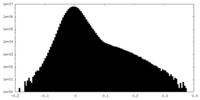
| Density Histograms |
- Sample components
Sample components
-Entire : rat GluN1a-2B Fab 003-102 local refinement
| Entire | Name: rat GluN1a-2B Fab 003-102 local refinement |
|---|---|
| Components |
|
-Supramolecule #1: rat GluN1a-2B Fab 003-102 local refinement
| Supramolecule | Name: rat GluN1a-2B Fab 003-102 local refinement / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glutamate receptor ionotropic, NMDA 1
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.352012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KIVNIGAVLS TRKHEQMFRE AVNQANKRHG SWKIQLQATS VTHKPNAIQM ALSVCEDLIS SQVYAILVSH PPTPNDHFTP TPVSYTAGF YRIPVLGLTT RMSIYSDKSI HLSFLRTVPP YSHQSSVWFE MMRVYNWNHI ILLVSDDHEG RAAQKRLETL L EERESKAE ...String: KIVNIGAVLS TRKHEQMFRE AVNQANKRHG SWKIQLQATS VTHKPNAIQM ALSVCEDLIS SQVYAILVSH PPTPNDHFTP TPVSYTAGF YRIPVLGLTT RMSIYSDKSI HLSFLRTVPP YSHQSSVWFE MMRVYNWNHI ILLVSDDHEG RAAQKRLETL L EERESKAE KVLQFDPGTK NVTALLMEAR ELEARVIILS ASEDDAATVY RAAAMLDMTG SGYVWLVGER EISGNALRYA PD GIIGLQL INGKNESAHI SDAVGVVAQA VHELLEKENI TDPPRGCVGN TNIWKTGPLF KRVLMSSKYA DGVTGRVEFN EDG DRKFAQ YSIMNLQNRK LVQVGIYNGT HVIPNDRKII WPGGETEKPR GYQ UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Glutamate receptor ionotropic, NMDA 2B
| Macromolecule | Name: Glutamate receptor ionotropic, NMDA 2B / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.103426 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SIGIAVILVG TSDEVAIKDA HEKDDFHHLS VVPRVELVAM NETDPKSIIT RICDLMSDRK IQGVVFADDT DQEAIAQILD FISAQTLTP ILGIHGGSSM IMADKDESSM FFQFGPSIEQ QASVMLNIME EYDWYIFSIV TTYFPGYQDF VNKIRSTIEN S FVGWELEE ...String: SIGIAVILVG TSDEVAIKDA HEKDDFHHLS VVPRVELVAM NETDPKSIIT RICDLMSDRK IQGVVFADDT DQEAIAQILD FISAQTLTP ILGIHGGSSM IMADKDESSM FFQFGPSIEQ QASVMLNIME EYDWYIFSIV TTYFPGYQDF VNKIRSTIEN S FVGWELEE VLLLDMSLDD GDSKIQNQLK KLQSPIILLY CTKEEATYIF EVANSVGLTG YGYTWIVPSL VAGDTDTVPS EF PTGLISV SYDEWDYGLP ARVRDGIAII TTAASDMLSE HSFIPEPKSS CYNTHEKRIY QSNMLNRYLI NVTFEGRDLS FSE EGYQMH PKLVIILLNK ERKWERVGKW KDKSLQMK UniProtKB: Glutamate receptor ionotropic, NMDA 2B |
-Macromolecule #3: 003-102 Heavy
| Macromolecule | Name: 003-102 Heavy / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.477862 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LQLQESGPGL VKPSQTLSLT CTVSGGSISS SNWWSWVRQP PGKGLEWIGE IYHSGNTNYN PSLKSRVTVS VDKSKNQFSL KLTSVTAAD TAVYYCARDV SGGVNWFDPW GQGTLV |
-Macromolecule #4: 003-102 Light
| Macromolecule | Name: 003-102 Light / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.582475 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NFMLTQPHSV SESPGKTVTI SCTRSSGSIA SNYVQWYQQR PGSAPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LKTEDEADY YCQSYDSSTV VFGGGTKLT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)