[English] 日本語
 Yorodumi
Yorodumi- EMDB-38635: Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline | |||||||||
 Map data Map data | refined map without post process | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / Tigecycline / antibiotic / CCDC124 | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial translational termination / mitochondrial translational elongation / translation release factor activity, codon nonspecific / Mitochondrial translation elongation / Mitochondrial translation termination / Mitochondrial translation initiation / translation release factor activity / mitochondrial large ribosomal subunit / peptidyl-tRNA hydrolase / mitochondrial ribosome ...mitochondrial translational termination / mitochondrial translational elongation / translation release factor activity, codon nonspecific / Mitochondrial translation elongation / Mitochondrial translation termination / Mitochondrial translation initiation / translation release factor activity / mitochondrial large ribosomal subunit / peptidyl-tRNA hydrolase / mitochondrial ribosome / Hydrolases; Acting on ester bonds; Endoribonucleases producing 5'-phosphomonoesters / mitochondrial small ribosomal subunit / peptidyl-tRNA hydrolase activity / mitochondrial translation / anatomical structure morphogenesis / RNA processing / rescue of stalled ribosome / Mitochondrial protein degradation / cellular response to leukemia inhibitory factor / fibrillar center / cell junction / large ribosomal subunit / double-stranded RNA binding / large ribosomal subunit rRNA binding / small ribosomal subunit rRNA binding / endonuclease activity / mitochondrial inner membrane / negative regulation of translation / rRNA binding / nuclear body / ribosome / structural constituent of ribosome / mitochondrial matrix / protein domain specific binding / translation / ribonucleoprotein complex / nucleotide binding / mRNA binding / apoptotic process / nucleolus / mitochondrion / RNA binding / nucleoplasm / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
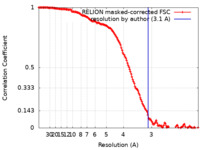
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Li X / Wang M / Cheng J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline. Authors: Xiang Li / Mengjiao Wang / Timo Denk / Robert Buschauer / Yi Li / Roland Beckmann / Jingdong Cheng /   Abstract: Tigecycline is widely used for treating complicated bacterial infections for which there are no effective drugs. It inhibits bacterial protein translation by blocking the ribosomal A-site. However, ...Tigecycline is widely used for treating complicated bacterial infections for which there are no effective drugs. It inhibits bacterial protein translation by blocking the ribosomal A-site. However, even though it is also cytotoxic for human cells, the molecular mechanism of its inhibition remains unclear. Here, we present cryo-EM structures of tigecycline-bound human mitochondrial 55S, 39S, cytoplasmic 80S and yeast cytoplasmic 80S ribosomes. We find that at clinically relevant concentrations, tigecycline effectively targets human 55S mitoribosomes, potentially, by hindering A-site tRNA accommodation and by blocking the peptidyl transfer center. In contrast, tigecycline does not bind to human 80S ribosomes under physiological concentrations. However, at high tigecycline concentrations, in addition to blocking the A-site, both human and yeast 80S ribosomes bind tigecycline at another conserved binding site restricting the movement of the L1 stalk. In conclusion, the observed distinct binding properties of tigecycline may guide new pathways for drug design and therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38635.map.gz emd_38635.map.gz | 224.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38635-v30.xml emd-38635-v30.xml emd-38635.xml emd-38635.xml | 72.6 KB 72.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38635_fsc.xml emd_38635_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_38635.png emd_38635.png | 121.5 KB | ||
| Filedesc metadata |  emd-38635.cif.gz emd-38635.cif.gz | 15.2 KB | ||
| Others |  emd_38635_additional_1.map.gz emd_38635_additional_1.map.gz emd_38635_half_map_1.map.gz emd_38635_half_map_1.map.gz emd_38635_half_map_2.map.gz emd_38635_half_map_2.map.gz | 161.2 MB 225.6 MB 225.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38635 http://ftp.pdbj.org/pub/emdb/structures/EMD-38635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38635 | HTTPS FTP |
-Validation report
| Summary document |  emd_38635_validation.pdf.gz emd_38635_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38635_full_validation.pdf.gz emd_38635_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_38635_validation.xml.gz emd_38635_validation.xml.gz | 22.7 KB | Display | |
| Data in CIF |  emd_38635_validation.cif.gz emd_38635_validation.cif.gz | 30 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38635 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38635 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38635 | HTTPS FTP |
-Related structure data
| Related structure data |  8xt3MC  8k2aC  8k2bC  8k2cC  8k2dC  8k82C  8xsxC  8xsyC  8xszC  8xt0C  8xt1C  8xt2C  8yooC  8yopC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38635.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38635.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined map without post process | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
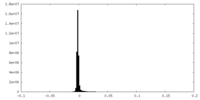
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
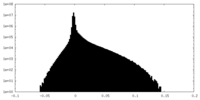
-Additional map: local resolution filtered map using Relion
| File | emd_38635_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map using Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
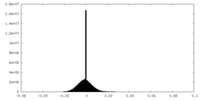
-Half map: #1
| File | emd_38635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
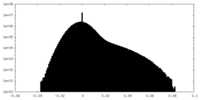
-Half map: #2
| File | emd_38635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 55S ribosome with tigecycline
+Supramolecule #1: 55S ribosome with tigecycline
+Macromolecule #1: 16s rRNA
+Macromolecule #2: Val tRNA
+Macromolecule #3: Large ribosomal subunit protein uL2m
+Macromolecule #4: Large ribosomal subunit protein uL3m
+Macromolecule #5: Large ribosomal subunit protein uL4m
+Macromolecule #6: Large ribosomal subunit protein bL9m
+Macromolecule #7: Large ribosomal subunit protein uL10m
+Macromolecule #8: Large ribosomal subunit protein uL11m
+Macromolecule #9: Large ribosomal subunit protein uL13m
+Macromolecule #10: Large ribosomal subunit protein uL14m
+Macromolecule #11: Large ribosomal subunit protein uL15m
+Macromolecule #12: Large ribosomal subunit protein uL16m
+Macromolecule #13: Large ribosomal subunit protein bL17m
+Macromolecule #14: Mitochondrial ribosomal protein L18, isoform CRA_b
+Macromolecule #15: Large ribosomal subunit protein bL19m
+Macromolecule #16: Large ribosomal subunit protein bL20m
+Macromolecule #17: Large ribosomal subunit protein bL21m
+Macromolecule #18: 39S ribosomal protein L22, mitochondrial
+Macromolecule #19: Large ribosomal subunit protein uL23m
+Macromolecule #20: Large ribosomal subunit protein uL24m
+Macromolecule #21: Large ribosomal subunit protein bL27m
+Macromolecule #22: Large ribosomal subunit protein bL28m
+Macromolecule #23: Large ribosomal subunit protein uL29m
+Macromolecule #24: Large ribosomal subunit protein uL30m
+Macromolecule #25: Large ribosomal subunit protein bL32m
+Macromolecule #26: Large ribosomal subunit protein bL33m
+Macromolecule #27: Large ribosomal subunit protein bL34m
+Macromolecule #28: Large ribosomal subunit protein bL35m
+Macromolecule #29: Large ribosomal subunit protein bL36m
+Macromolecule #30: Large ribosomal subunit protein mL37
+Macromolecule #31: Large ribosomal subunit protein mL38
+Macromolecule #32: Large ribosomal subunit protein mL39
+Macromolecule #33: Large ribosomal subunit protein mL40
+Macromolecule #34: Large ribosomal subunit protein mL41
+Macromolecule #35: Large ribosomal subunit protein mL42
+Macromolecule #36: Large ribosomal subunit protein mL43
+Macromolecule #37: Large ribosomal subunit protein mL44
+Macromolecule #38: Large ribosomal subunit protein mL45
+Macromolecule #39: Large ribosomal subunit protein mL46
+Macromolecule #40: Large ribosomal subunit protein mL48
+Macromolecule #41: Large ribosomal subunit protein mL49
+Macromolecule #42: Large ribosomal subunit protein mL50
+Macromolecule #43: Large ribosomal subunit protein mL51
+Macromolecule #44: 39S ribosomal protein L52, mitochondrial
+Macromolecule #45: Large ribosomal subunit protein mL53
+Macromolecule #46: Large ribosomal subunit protein mL54
+Macromolecule #47: Large ribosomal subunit protein mL55
+Macromolecule #48: Large ribosomal subunit protein mL63
+Macromolecule #49: Large ribosomal subunit protein mL62
+Macromolecule #50: Large ribosomal subunit protein mL64
+Macromolecule #51: Large ribosomal subunit protein mL66
+Macromolecule #52: Large ribosomal subunit protein mL65
+Macromolecule #53: MAGNESIUM ION
+Macromolecule #54: TIGECYCLINE
+Macromolecule #55: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)