[English] 日本語
 Yorodumi
Yorodumi- EMDB-38639: Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecyc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecycline (focusing on 28S ribosome head) | |||||||||
 Map data Map data | refined map from multibody refinement focusing on 28S mitoribosome head | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / Tigecycline / antibiotic / CCDC124 | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Li X / Wang M / Cheng J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline. Authors: Xiang Li / Mengjiao Wang / Timo Denk / Robert Buschauer / Yi Li / Roland Beckmann / Jingdong Cheng /   Abstract: Tigecycline is widely used for treating complicated bacterial infections for which there are no effective drugs. It inhibits bacterial protein translation by blocking the ribosomal A-site. However, ...Tigecycline is widely used for treating complicated bacterial infections for which there are no effective drugs. It inhibits bacterial protein translation by blocking the ribosomal A-site. However, even though it is also cytotoxic for human cells, the molecular mechanism of its inhibition remains unclear. Here, we present cryo-EM structures of tigecycline-bound human mitochondrial 55S, 39S, cytoplasmic 80S and yeast cytoplasmic 80S ribosomes. We find that at clinically relevant concentrations, tigecycline effectively targets human 55S mitoribosomes, potentially, by hindering A-site tRNA accommodation and by blocking the peptidyl transfer center. In contrast, tigecycline does not bind to human 80S ribosomes under physiological concentrations. However, at high tigecycline concentrations, in addition to blocking the A-site, both human and yeast 80S ribosomes bind tigecycline at another conserved binding site restricting the movement of the L1 stalk. In conclusion, the observed distinct binding properties of tigecycline may guide new pathways for drug design and therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38639.map.gz emd_38639.map.gz | 148 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38639-v30.xml emd-38639-v30.xml emd-38639.xml emd-38639.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38639.png emd_38639.png | 93.4 KB | ||
| Filedesc metadata |  emd-38639.cif.gz emd-38639.cif.gz | 4.1 KB | ||
| Others |  emd_38639_additional_1.map.gz emd_38639_additional_1.map.gz emd_38639_half_map_1.map.gz emd_38639_half_map_1.map.gz emd_38639_half_map_2.map.gz emd_38639_half_map_2.map.gz | 156.3 MB 142.9 MB 142.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38639 http://ftp.pdbj.org/pub/emdb/structures/EMD-38639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38639 | HTTPS FTP |
-Related structure data
| Related structure data |  8k2aC  8k2bC  8k2cC  8k2dC  8k82C  8xsxC  8xsyC  8xszC  8xt0C  8xt1C  8xt2C  8xt3C  8yooC  8yopC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38639.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38639.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined map from multibody refinement focusing on 28S mitoribosome head | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
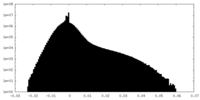
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: local resolution filtered map using Relion
| File | emd_38639_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map using Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
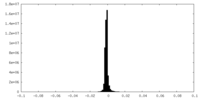
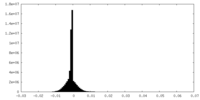
| Density Histograms |
-Half map: #1
| File | emd_38639_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
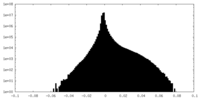
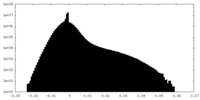
| Density Histograms |
-Half map: #2
| File | emd_38639_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
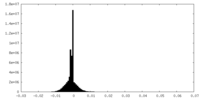
| Density Histograms |
- Sample components
Sample components
-Entire : 55S ribosome with tigecycline
| Entire | Name: 55S ribosome with tigecycline |
|---|---|
| Components |
|
-Supramolecule #1: 55S ribosome with tigecycline
| Supramolecule | Name: 55S ribosome with tigecycline / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#86 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Relion |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 83274 |
| Initial angle assignment | Type: OTHER / Software - Name: RELION / Details: Relion |
| Final angle assignment | Type: OTHER / Software - Name: RELION / Details: Relion |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)