[English] 日本語
 Yorodumi
Yorodumi- EMDB-12563: Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiqu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12563 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GID / CTLH / ubiquitin / E3 ligase / supramolecular assembly / metabolism / gluconeogenesis / cryoEM / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationGID complex / Regulation of pyruvate metabolism / traversing start control point of mitotic cell cycle / regulation of nitrogen utilization / negative regulation of gluconeogenesis / cytoskeleton organization / peroxisome / proteasome-mediated ubiquitin-dependent protein catabolic process / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
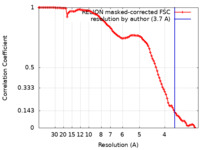
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Chrustowicz J / Sherpa D / Prabu JR / Schulman BA | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme. Authors: Dawafuti Sherpa / Jakub Chrustowicz / Shuai Qiao / Christine R Langlois / Laura A Hehl / Karthik Varma Gottemukkala / Fynn M Hansen / Ozge Karayel / Susanne von Gronau / J Rajan Prabu / ...Authors: Dawafuti Sherpa / Jakub Chrustowicz / Shuai Qiao / Christine R Langlois / Laura A Hehl / Karthik Varma Gottemukkala / Fynn M Hansen / Ozge Karayel / Susanne von Gronau / J Rajan Prabu / Matthias Mann / Arno F Alpi / Brenda A Schulman /  Abstract: How are E3 ubiquitin ligases configured to match substrate quaternary structures? Here, by studying the yeast GID complex (mutation of which causes deficiency in glucose-induced degradation of ...How are E3 ubiquitin ligases configured to match substrate quaternary structures? Here, by studying the yeast GID complex (mutation of which causes deficiency in glucose-induced degradation of gluconeogenic enzymes), we discover supramolecular chelate assembly as an E3 ligase strategy for targeting an oligomeric substrate. Cryoelectron microscopy (cryo-EM) structures show that, to bind the tetrameric substrate fructose-1,6-bisphosphatase (Fbp1), two minimally functional GID E3s assemble into the 20-protein Chelator-GID, which resembles an organometallic supramolecular chelate. The Chelator-GID assembly avidly binds multiple Fbp1 degrons so that multiple Fbp1 protomers are simultaneously ubiquitylated at lysines near the allosteric and substrate binding sites. Importantly, key structural and biochemical features, including capacity for supramolecular assembly, are preserved in the human ortholog, the CTLH E3. Based on our integrative structural, biochemical, and cell biological data, we propose that higher-order E3 ligase assembly generally enables multipronged targeting, capable of simultaneously incapacitating multiple protomers and functionalities of oligomeric substrates. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme Authors: Sherpa D / Chrustowicz J / Qiao S / Langlois CR / Hehl LA / Gottemukkala KV / Hansen FM / Karayel O / Prabu JR / Mann M / Alpi AF / Schulman BA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12563.map.gz emd_12563.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12563-v30.xml emd-12563-v30.xml emd-12563.xml emd-12563.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12563_fsc.xml emd_12563_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12563.png emd_12563.png | 130.6 KB | ||
| Masks |  emd_12563_msk_1.map emd_12563_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12563.cif.gz emd-12563.cif.gz | 7.1 KB | ||
| Others |  emd_12563_additional_1.map.gz emd_12563_additional_1.map.gz emd_12563_half_map_1.map.gz emd_12563_half_map_1.map.gz emd_12563_half_map_2.map.gz emd_12563_half_map_2.map.gz | 81 MB 71.3 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12563 http://ftp.pdbj.org/pub/emdb/structures/EMD-12563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12563 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12563 | HTTPS FTP |
-Validation report
| Summary document |  emd_12563_validation.pdf.gz emd_12563_validation.pdf.gz | 682.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12563_full_validation.pdf.gz emd_12563_full_validation.pdf.gz | 682.1 KB | Display | |
| Data in XML |  emd_12563_validation.xml.gz emd_12563_validation.xml.gz | 17.3 KB | Display | |
| Data in CIF |  emd_12563_validation.cif.gz emd_12563_validation.cif.gz | 22.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12563 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12563 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12563 | HTTPS FTP |
-Related structure data
| Related structure data |  7nsbMC  7ns3C  7ns4C  7ns5C  7nscC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12563.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12563.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.58 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12563_msk_1.map emd_12563_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_12563_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12563_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12563_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Supramolecular assembly module of yeast Chelator-GID SR4 comprisi...
| Entire | Name: Supramolecular assembly module of yeast Chelator-GID SR4 comprising Gid1, Gid8 and a homodimer of Gid7 |
|---|---|
| Components |
|
-Supramolecule #1: Supramolecular assembly module of yeast Chelator-GID SR4 comprisi...
| Supramolecule | Name: Supramolecular assembly module of yeast Chelator-GID SR4 comprising Gid1, Gid8 and a homodimer of Gid7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Generated by focused refinement of Chelator-GID SR4 + Fbp1 map |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 330 KDa |
-Macromolecule #1: Vacuolar import and degradation protein 30
| Macromolecule | Name: Vacuolar import and degradation protein 30 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 108.28768 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSEYMDDVDR EFINCLFPSY LLQQPVAYDL WILYLQHRKL FHKLKNTNLI NADENPTGVG MGRTKLTALT RKEIWSKLMN LGVLGTISF EAVNDDYLIQ VYKYFYPDVN DFTLRFGVKD SNKNSVRVMK ASSDMRKNAQ ELLEPVLSER EMALNSNTSL E NDRNDDDD ...String: MSEYMDDVDR EFINCLFPSY LLQQPVAYDL WILYLQHRKL FHKLKNTNLI NADENPTGVG MGRTKLTALT RKEIWSKLMN LGVLGTISF EAVNDDYLIQ VYKYFYPDVN DFTLRFGVKD SNKNSVRVMK ASSDMRKNAQ ELLEPVLSER EMALNSNTSL E NDRNDDDD DDDDDDDDDD DDDDDDDESD LESLEGEVDT DTDDNNEGDG SDNHEEGGEE GSRGADADVS SAQQRAERVA DP WIYQRSR SAINIETESR NLWDTSDKNS GLQYYPPDQS PSSSFSSPRV SSGNDKNDNE ATNVLSNSGS KKKNSMIPDI YKI LGYFLP SRWQAQPNNS LQLSQDGITH LQPNPDYHSY MTYERSSASS ASTRNRLRTS FENSGKVDFA VTWANKSLPD NKLT IFYYE IKVLSVTSTE SAENSNIVIG YKLVENELME ATTKKSVSRS SVAGSSSSLG GSNNMSSNRV PSTSFTMEGT QRRDY IYEG GVSAMSLNVD GSINKCQKYG FDLNVFGYCG FDGLITNSTE QSKEYAKPFG RDDVIGCGIN FIDGSIFFTK NGIHLG NAF TDLNDLEFVP YVALRPGNSI KTNFGLNEDF VFDIIGYQDK WKSLAYEHIC RGRQMDVSIE EFDSDESEED ETENGPE EN KSTNVNEDLM DIDQEDGAAG NKDTKKLNDE KDNNLKFLLG EDNRFIDGKL VRPDVNNINN LSVDDGSLPN TLNVMIND Y LIHEGLVDVA KGFLKDLQKD AVNVNGQHSE SKDVIRHNER QIMKEERMVK IRQELRYLIN KGQISKCINY IDNEIPDLL KNNLELVFEL KLANYLVMIK KSSSKDDDEI ENLILKGQEL SNEFIYDTKI PQSLRDRFSG QLSNVSALLA YSNPLVEAPK EISGYLSDE YLQERLFQVS NNTILTFLHK DSECALENVI SNTRAMLSTL LEYNAFGSTN SSDPRYYKAI NFDEDVLNL UniProtKB: Vacuolar import and degradation protein 30 |
-Macromolecule #2: Glucose-induced degradation protein 7
| Macromolecule | Name: Glucose-induced degradation protein 7 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 84.607297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSHTNKIAYV LNNDTEETAS PSSVGCFDKK QLTKLLIHTL KELGYDSAAN QLLLESGGYQ NESNHIQTFF KLIKTGQFHL INWQIVCSL PLAHSSPLRS EWLQRLLIPT PTPATTSLFD HMLLQLQYLQ QLMSSVNSST CSDAEIATLR NYVEIMILVN R QIFLEFFH ...String: MSHTNKIAYV LNNDTEETAS PSSVGCFDKK QLTKLLIHTL KELGYDSAAN QLLLESGGYQ NESNHIQTFF KLIKTGQFHL INWQIVCSL PLAHSSPLRS EWLQRLLIPT PTPATTSLFD HMLLQLQYLQ QLMSSVNSST CSDAEIATLR NYVEIMILVN R QIFLEFFH PVTNSASHKG PHTALPVLYL RKILKNFIEI WDSLLVSNDQ FLNEENIFNP ETTLRELSTY LTNPKLTAQL NL ERDHLID AISKYIDPNE LVPKGRLLHL LKQAIKYQQS QDIFNIIDPD DDASFSSPPH RINLLQDNFS HDLTVTFQEW KTI QDTTDE IWFLTFSPNG KYLASATSES SRGYFITVYD VEQDFKIYKT CVSLSQSVLY LMFSPDSRYL VACPFSEDVT IYDM NATSL PDASATDSFL LYPSTRLSPM DSFKLDTTTY PDDTESSASS SSRPANANSN QSRVWCCDAF HTAERAGWMV VGSPD REAI VHSLTTKESL FSLKGRTCIA LGHDENISGR KSIDPAKVLY KPTSSNGNWQ YVEDDETFPR VHDVKISYDD KYVLLM THQ GVIDVYDFSG FPSKEELSKQ TVDPKNFLIP RIARLDVGKN MTCISLPLNT THQGFHRQQI SESQHLVLVS LQDNELQ MW DYKENILIQK YFGQKQQHFI IRSCFAYGNK LVMSGSEDGK IYIWDRIRGN LVSVLSGHST VMSNSTKPMG KNCNVVAS N PADKEMFASG GDDGKIKIWK ISRN UniProtKB: Glucose-induced degradation protein 7 |
-Macromolecule #3: Glucose-induced degradation protein 8
| Macromolecule | Name: Glucose-induced degradation protein 8 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 55.803309 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MTISTLSNET TKSGSCSGQG KNGGKDFTYG KKCFTKEEWK EQVAKYSAMG ELYANKTIHY PLKIQPNSSG GSQDEGFATI QTTPIEPTL PRLLLNYFVS MAYEDSSIRM AKELGFIRNN KDIAVFNDLY KIKERFHIKH LIKLGRINEA MEEINSIFGL E VLEETFNA ...String: MTISTLSNET TKSGSCSGQG KNGGKDFTYG KKCFTKEEWK EQVAKYSAMG ELYANKTIHY PLKIQPNSSG GSQDEGFATI QTTPIEPTL PRLLLNYFVS MAYEDSSIRM AKELGFIRNN KDIAVFNDLY KIKERFHIKH LIKLGRINEA MEEINSIFGL E VLEETFNA TGSYTGRTDR QQQQQQQQFD IDGDLHFKLL LLNLIEMIRS HHQQENITKD SNDFILNLIQ YSQNKLAIKA SS SVKKMQE LELAMTLLLF PLSDSADSGS IKLPKSLQNL YSISLRSKIA DLVNEKLLKF IHPRIQFEIS NNNSKFPDLL NSD KKIITQ NFTVYNNNLV NGSNGTKITH ISSDQPINEK MSSNEVTAAA NSVWLNQRDG NVGTGSAATT FHNLENKNYW NQTS ELLSS SNGKEKGLEF NNYYSSEFPY EPRLTQIMKL WCWCENQLHH NQIGVPRVEN SDENLYFQSG WSHPQFEKGG GSGGG SGGS AWSHPQFEK UniProtKB: Glucose-induced degradation protein 8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 79.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)