[English] 日本語
 Yorodumi
Yorodumi- EMDB-11583: Outer Dynein Arm-Shulin complex - Dyh5 motor (Tetrahymena thermophila) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11583 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer Dynein Arm-Shulin complex - Dyh5 motor (Tetrahymena thermophila) | |||||||||
 Map data Map data | Main map of Dyh5 motor (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -148 angstrom auto-sharpened map. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
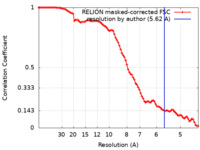
| Method | single particle reconstruction / cryo EM / Resolution: 5.62 Å | |||||||||
 Authors Authors | Mali GR / Abid Ali F / Lau CK / Carter AP | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Shulin packages axonemal outer dynein arms for ciliary targeting. Authors: Girish R Mali / Ferdos Abid Ali / Clinton K Lau / Farida Begum / Jérôme Boulanger / Jonathan D Howe / Zhuo A Chen / Juri Rappsilber / Mark Skehel / Andrew P Carter /   Abstract: The main force generators in eukaryotic cilia and flagella are axonemal outer dynein arms (ODAs). During ciliogenesis, these ~1.8-megadalton complexes are assembled in the cytoplasm and targeted to ...The main force generators in eukaryotic cilia and flagella are axonemal outer dynein arms (ODAs). During ciliogenesis, these ~1.8-megadalton complexes are assembled in the cytoplasm and targeted to cilia by an unknown mechanism. Here, we used the ciliate to identify two factors (Q22YU3 and Q22MS1) that bind ODAs in the cytoplasm and are required for ODA delivery to cilia. Q22YU3, which we named Shulin, locked the ODA motor domains into a closed conformation and inhibited motor activity. Cryo-electron microscopy revealed how Shulin stabilized this compact form of ODAs by binding to the dynein tails. Our findings provide a molecular explanation for how newly assembled dyneins are packaged for delivery to the cilia. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11583.map.gz emd_11583.map.gz | 28.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11583-v30.xml emd-11583-v30.xml emd-11583.xml emd-11583.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11583_fsc.xml emd_11583_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11583.png emd_11583.png | 57.2 KB | ||
| Masks |  emd_11583_msk_1.map emd_11583_msk_1.map | 43.5 MB |  Mask map Mask map | |
| Others |  emd_11583_additional_1.map.gz emd_11583_additional_1.map.gz emd_11583_half_map_1.map.gz emd_11583_half_map_1.map.gz emd_11583_half_map_2.map.gz emd_11583_half_map_2.map.gz | 173.7 KB 50 MB 50 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11583 http://ftp.pdbj.org/pub/emdb/structures/EMD-11583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11583 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11583 | HTTPS FTP |
-Validation report
| Summary document |  emd_11583_validation.pdf.gz emd_11583_validation.pdf.gz | 397.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11583_full_validation.pdf.gz emd_11583_full_validation.pdf.gz | 396.9 KB | Display | |
| Data in XML |  emd_11583_validation.xml.gz emd_11583_validation.xml.gz | 14 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11583 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11583 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11583 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11583 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11583.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11583.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map of Dyh5 motor (resampled on a full-length ODA-Shulin structure from a smaller subset of particles using EMDA). -148 angstrom auto-sharpened map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.22991 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11583_msk_1.map emd_11583_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Mask for Dyh5 motor map (Z-flipped, non resampled)...
| File | emd_11583_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mask for Dyh5 motor map (Z-flipped, non resampled) to be used with half maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of Dyh5 motor map (Z-flipped, non-resampled)
| File | emd_11583_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of Dyh5 motor map (Z-flipped, non-resampled) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of Dyh5 motor map (Z-flipped, non-resampled)
| File | emd_11583_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of Dyh5 motor map (Z-flipped, non-resampled) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrahymena thermophila ODA Dyh5 motor
| Entire | Name: Tetrahymena thermophila ODA Dyh5 motor |
|---|---|
| Components |
|
-Supramolecule #1: Tetrahymena thermophila ODA Dyh5 motor
| Supramolecule | Name: Tetrahymena thermophila ODA Dyh5 motor / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)