[English] 日本語
 Yorodumi
Yorodumi- EMDB-11269: E2 core of the fungal Pyruvate dehydrogenase complex with absent ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11269 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E2 core of the fungal Pyruvate dehydrogenase complex with absent periphery, structured core and empty interior. | ||||||||||||||||||
 Map data Map data | Fungal PDC (N. crassa). Recombinant preparation-E2. Enforced symmetry: I2. Periphery (E2 only) is flexible/oversym. Core is structured. Interior is empty. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Neurospora crassa (fungus) Neurospora crassa (fungus) | ||||||||||||||||||
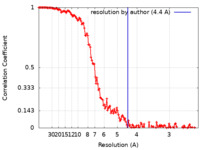
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||||||||
 Authors Authors | Forsberg BO / Lindahl E / Aibara S / Howard RJ / Mortezaei N | ||||||||||||||||||
| Funding support |  Sweden, European Union, 5 items Sweden, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex. Authors: B O Forsberg / S Aibara / R J Howard / N Mortezaei / E Lindahl /   Abstract: The pyruvate dehydrogenase complex (PDC) is a multienzyme complex central to aerobic respiration, connecting glycolysis to mitochondrial oxidation of pyruvate. Similar to the E3-binding protein (E3BP) ...The pyruvate dehydrogenase complex (PDC) is a multienzyme complex central to aerobic respiration, connecting glycolysis to mitochondrial oxidation of pyruvate. Similar to the E3-binding protein (E3BP) of mammalian PDC, PX selectively recruits E3 to the fungal PDC, but its divergent sequence suggests a distinct structural mechanism. Here, we report reconstructions of PDC from the filamentous fungus Neurospora crassa by cryo-electron microscopy, where we find protein X (PX) interior to the PDC core as opposed to substituting E2 core subunits as in mammals. Steric occlusion limits PX binding, resulting in predominantly tetrahedral symmetry, explaining previous observations in Saccharomyces cerevisiae. The PX-binding site is conserved in (and specific to) fungi, and complements possible C-terminal binding motifs in PX that are absent in mammalian E3BP. Consideration of multiple symmetries thus reveals a differential structural basis for E3BP-like function in fungal PDC. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11269.map.gz emd_11269.map.gz | 28.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11269-v30.xml emd-11269-v30.xml emd-11269.xml emd-11269.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11269_fsc.xml emd_11269_fsc.xml | 17.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_11269.png emd_11269.png | 93.8 KB | ||
| Others |  emd_11269_half_map_1.map.gz emd_11269_half_map_1.map.gz emd_11269_half_map_2.map.gz emd_11269_half_map_2.map.gz | 170.1 MB 170.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11269 http://ftp.pdbj.org/pub/emdb/structures/EMD-11269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11269 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11269 | HTTPS FTP |
-Validation report
| Summary document |  emd_11269_validation.pdf.gz emd_11269_validation.pdf.gz | 325.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11269_full_validation.pdf.gz emd_11269_full_validation.pdf.gz | 324.6 KB | Display | |
| Data in XML |  emd_11269_validation.xml.gz emd_11269_validation.xml.gz | 19.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11269 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11269 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11269 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11269.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11269.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fungal PDC (N. crassa). Recombinant preparation-E2. Enforced symmetry: I2. Periphery (E2 only) is flexible/oversym. Core is structured. Interior is empty. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half-map 1. Fungal PDC (N. crassa). Recombinant preparation-E2....
| File | emd_11269_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1. Fungal PDC (N. crassa). Recombinant preparation-E2. Enforced symmetry: I2. Periphery (E2 only) is flexible/oversym. Core is structured. Interior is empty. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2. Fungal PDC (N. crassa). Recombinant preparation-E2....
| File | emd_11269_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2. Fungal PDC (N. crassa). Recombinant preparation-E2. Enforced symmetry: I2. Periphery (E2 only) is flexible/oversym. Core is structured. Interior is empty. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E2 core of the fungal Pyruvate dehydrogenase complex with absent ...
| Entire | Name: E2 core of the fungal Pyruvate dehydrogenase complex with absent periphery, structured core and empty interior. |
|---|---|
| Components |
|
-Supramolecule #1: E2 core of the fungal Pyruvate dehydrogenase complex with absent ...
| Supramolecule | Name: E2 core of the fungal Pyruvate dehydrogenase complex with absent periphery, structured core and empty interior. type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 33.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)