3B9L
 
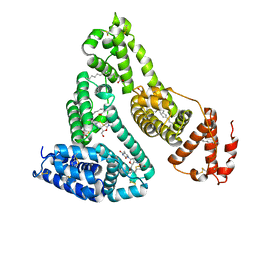 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3B9M
 
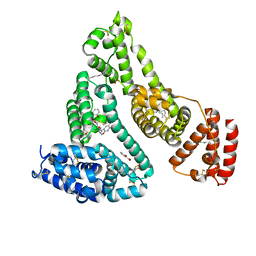 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
104D
 
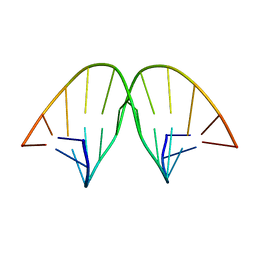 | |
1ZHU
 
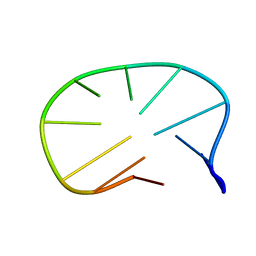 | | DNA (5'-D(*CP*AP*AP*TP*GP*CP*AP*AP*TP*G)-3'), NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*AP*TP*GP*CP*AP*AP*TP*G)-3') | | Authors: | Zhu, L, Chou, S.-H, Xu, J, Reid, B.R. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a single-cytidine hairpin loop formed by the DNA triplet GCA.
Nat.Struct.Biol., 2, 1995
|
|
1LPV
 
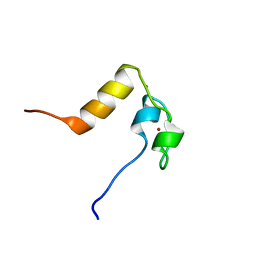 | | DROSOPHILA MELANOGASTER DOUBLESEX (DSX), NMR, 18 STRUCTURES | | Descriptor: | Doublesex protein, ZINC ION | | Authors: | Zhu, L, Wilken, J, Phillips, N, Narendra, U, Chan, G, Stratton, S, Kent, S, Weiss, M.A. | | Deposit date: | 2002-05-08 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers.
Genes Dev., 14, 2000
|
|
1XUE
 
 | |
3JB4
 
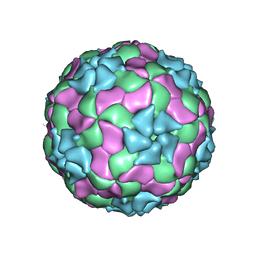 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
4RR4
 
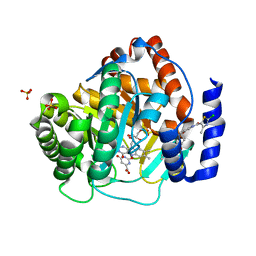 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367 | | Descriptor: | 2-chloro-N-[3-(4-{[(2Z)-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enoyl]amino}phenoxy)phenyl]-4-methyl-1,3-thiazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-11-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A367
TO BE PUBLISHED
|
|
4RKA
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347 | | Descriptor: | 2-{[5-(naphthalen-1-ylmethyl)-4-oxo-4H-1lambda~4~,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A347
TO BE PUBLISHED
|
|
4RK8
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356 | | Descriptor: | 5-fluoro-2-{[(5Z)-5-(naphthalen-1-ylmethylidene)-4-oxo-4,5-dihydro-1,3-thiazol-2-yl]amino}benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A356
TO BE PUBLISHED
|
|
4RLI
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhu, L, Ren, X, Zhu, J, Li, H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A048
TO BE PUBLISHED
|
|
4LS1
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312 | | Descriptor: | 2-[(E)-{2-[4-(2-chlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}methyl]benzoic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312
To be Published
|
|
4LS2
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A313 | | Descriptor: | 2-[(E)-{2-[4-(3-methoxyphenyl)-1,3-thiazol-2-yl]hydrazinylidene}methyl]benzoic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A313
To be Published
|
|
4LS0
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH01B0033 | | Descriptor: | 2-{(E)-[2-(4-phenyl-1,3-thiazol-2-yl)hydrazinylidene]methyl}benzaldehyde, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DHO1B0033
To be Published
|
|
4JGD
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016 | | Descriptor: | 1-[4-methyl-2-(naphthalen-2-ylamino)-1,3-thiazol-5-yl]ethanone, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016
TO BE PUBLISHED
|
|
4JTT
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 066 | | Descriptor: | 2,4-dimethyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4JTU
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with brequinar analogue | | Descriptor: | 6-bromo-2-{4-[(2R)-butan-2-yl]phenyl}-3-methylquinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4JS3
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057 | | Descriptor: | 2-chloro-4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4JTS
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 072 | | Descriptor: | 4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
6U4N
 
 | |
6U4M
 
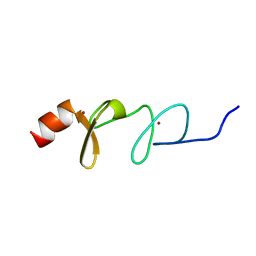 | | Solution structure of paxillin LIM4 | | Descriptor: | Paxillin, ZINC ION | | Authors: | Zhu, L, Qin, J. | | Deposit date: | 2019-08-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Paxillin Recruitment by Kindlin-2 in Regulating Cell Adhesion.
Structure, 27, 2019
|
|
6AKS
 
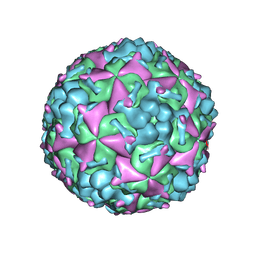 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
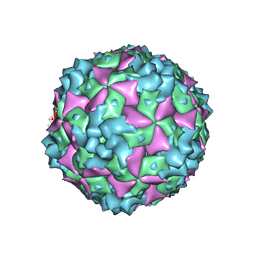 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
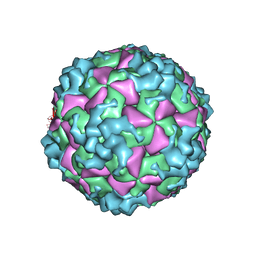 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
5GKA
 
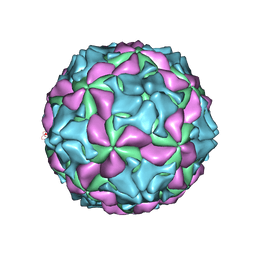 | | cryo-EM structure of human Aichi virus | | Descriptor: | Genome polyprotein, capsid protein VP0, capsid protein VP1 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Tuthill, T.J, Fry, E.E, Rao, Z.H, Stuart, D.I. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of human Aichi virus and implications for receptor binding
Nat Microbiol, 1, 2016
|
|
