1G5X
 
 | | The Structure of Beta-Ketoacyl-[Acyl Carrier Protein] Synthase I | | Descriptor: | BETA-KETOACYL ACYL CARRIER PROTEIN SYNTHASE I | | Authors: | Zhang, Y.M, Rao, M.S, Heath, R.J, Price, A.C, Olson, A.J, Rock, C.O, White, S.W. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III.
J.Biol.Chem., 276, 2001
|
|
2ALM
 
 | | Crystal structure analysis of a mutant beta-ketoacyl-[acyl carrier protein] synthase II from Streptococcus pneumoniae | | Descriptor: | 3-oxoacyl-(acyl-carrier-protein) synthase II, MAGNESIUM ION | | Authors: | Zhang, Y.M, Hurlbert, J, White, S.W, Rock, C.O. | | Deposit date: | 2005-08-07 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae.
J.Biol.Chem., 281, 2006
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
1NYK
 
 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
2AQB
 
 | | Structure-activity relationships at the 5-position of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherchia coli | | Descriptor: | (5R)-5-[(1E)-BUTA-1,3-DIENYL]-4-HYDROXY-3,5-DIMETHYLTHIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
2AQ7
 
 | | Structure-activity relationships at the 5-posiiton of thiolactomycin: an intact 5(R)-isoprene unit is required for activity against the condensing enzymes from Mycobacterium tuberculosis and Escherichia coli | | Descriptor: | (5R)-4-HYDROXY-3,5-DIMETHYL-5-[(1E,3E)-2-METHYLPENTA-1,3-DIENYL]THIOPHEN-2(5H)-ONE, 3-oxoacyl-[acyl-carrier-protein] synthase I | | Authors: | Kim, P, Zhang, Y.M, Shenoy, G, Nguyen, Q.A, Boshoff, H.I, Manjunatha, U.H, Goodwin, M.B, Lonsdale, J, Price, A.C, Miller, D.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationships at the 5-Position of Thiolactomycin: An Intact (5R)-Isoprene Unit Is Required for Activity against the Condensing Enzymes from Mycobacterium tuberculosis and Escherichia coli
J.Med.Chem., 49, 2006
|
|
2GQD
 
 | |
8TWP
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, R416A | | Descriptor: | Nucleoprotein | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
8TWR
 
 | | Influenza A virus (A/Aichi/2/1968(H3N2) nucleoprotein mutant - 2-7 deleted, P283S, R416A | | Descriptor: | Nucleoprotein, SODIUM ION | | Authors: | Yoon, J, Zhang, Y.M, Grant, R.A, Shoulders, M.D. | | Deposit date: | 2023-08-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The immune-evasive proline-283 substitution in influenza nucleoprotein increases aggregation propensity without altering the native structure.
Sci Adv, 10, 2024
|
|
7CQ7
 
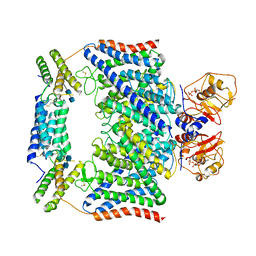 | | Structure of the human CLCN7-OSTM1 complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ADP
To Be Published
|
|
7CQ5
 
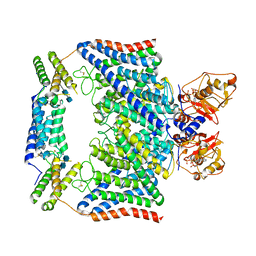 | | Structure of the human CLCN7-OSTM1 complex with ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ATP
To Be Published
|
|
7CQ6
 
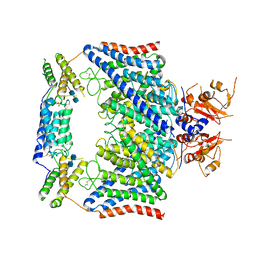 | | Structure of the human CLCN7-OSTM1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, H(+)/Cl(-) exchange transporter 7, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex
To Be Published
|
|
2K92
 
 | |
1KLP
 
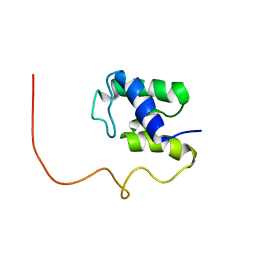 | | The Solution Structure of Acyl Carrier Protein from Mycobacterium tuberculosis | | Descriptor: | MEROMYCOLATE EXTENSION ACYL CARRIER PROTEIN | | Authors: | Wong, H.C, Liu, G, Zhang, Y.-M, Rock, C.O, Zheng, J. | | Deposit date: | 2001-12-12 | | Release date: | 2002-06-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of acyl carrier protein from Mycobacterium tuberculosis.
J.Biol.Chem., 277, 2002
|
|
3LSJ
 
 | |
3LSR
 
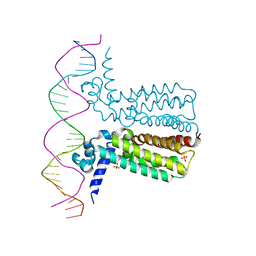 | |
3LSP
 
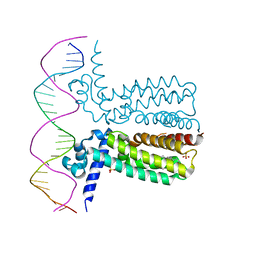 | | Crystal Structure of DesT bound to desCB promoter and oleoyl-CoA | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*TP*CP*GP*AP*GP*TP*CP*AP*AP*CP*AP*AP*GP*CP*GP*TP*TP*CP*AP*CP*TP*GP*AP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*TP*CP*AP*GP*TP*GP*AP*AP*CP*GP*CP*TP*TP*GP*TP*TP*GP*AP*CP*TP*CP*GP*AP*TP*TP*G)-3'), DesT, ... | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the transcriptional regulation of membrane lipid homeostasis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MK6
 
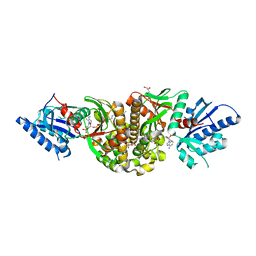 | | Substrate and Inhibitor Binding to Pank | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Pantothenate kinase 3 | | Authors: | Yun, M.-K, White, S.W. | | Deposit date: | 2010-04-14 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Modulation of Pantothenate Kinase 3 Activity by Small Molecules that Interact with the Substrate/Allosteric Regulatory Domain.
Chem.Biol., 17, 2010
|
|
1I01
 
 | |
1JYL
 
 | |
1JYK
 
 | |
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
2F9T
 
 | | Structure of the type III CoaA from Pseudomonas aeruginosa | | Descriptor: | Pantothenate Kinase | | Authors: | Leonardi, R, Yun, M.K, Chohnan, S, White, S.W, Rock, C.O, Jackowski, S. | | Deposit date: | 2005-12-06 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties.
Structure, 14, 2006
|
|
2EWS
 
 | |
8JGF
 
 | | CryoEM structure of Gq-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
