4M02
 
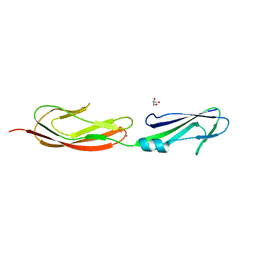 | | Middle fragment(residues 494-663) of the binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M03
 
 | | C-terminal fragment(residues 576-751) of binding region of SraP | | Descriptor: | CALCIUM ION, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M01
 
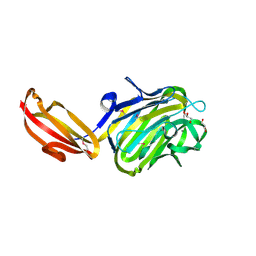 | | N terminal fragment(residues 245-575) of binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M00
 
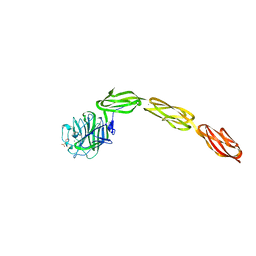 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4NT9
 
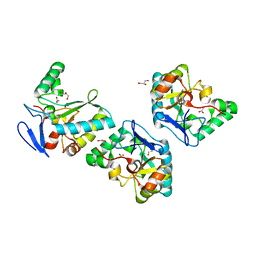 | | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia | | Descriptor: | ACETATE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yang, Y.H, Zhang, J, Jiang, Y.L, Zhou, C.Z, Chen, Y. | | Deposit date: | 2013-12-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia
To be Published
|
|
4PQG
 
 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
3PPR
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPP
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein, TRIMETHYL GLYCINE | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPO
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPN
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPQ
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
6K2I
 
 | |
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6JPM
 
 | |
6IYN
 
 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | Descriptor: | Nb26 | | Authors: | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
