1L8Z
 
 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
1L8Y
 
 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
1GDT
 
 | |
1T6W
 
 | | RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, hypothetical protein XP_346638 | | Authors: | Yang, W, Wilkins, A.L, Ye, Y, Liu, Z.-R, Urbauer, J.L, Kearney, A, van der Merwe, P.A, Yang, J.J. | | Deposit date: | 2004-05-07 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design of a calcium-binding protein with desired structure in a cell adhesion molecule.
J.Am.Chem.Soc., 127, 2005
|
|
1RNH
 
 | | STRUCTURE OF RIBONUCLEASE H PHASED AT 2 ANGSTROMS RESOLUTION BY MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | RIBONUCLEASE HI, SULFATE ION | | Authors: | Yang, W, Hendrickson, W.A, Crouch, R.J, Satow, Y. | | Deposit date: | 1990-07-11 | | Release date: | 1991-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein.
Science, 249, 1990
|
|
3LXF
 
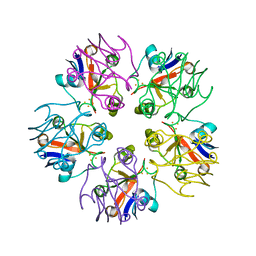 | | Crystal Structure of [2Fe-2S] Ferredoxin Arx from Novosphingobium aromaticivorans | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXI
 
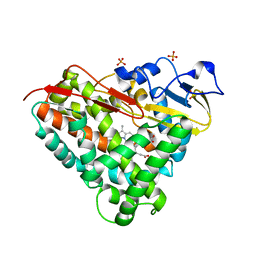 | | Crystal Structure of Camphor-Bound CYP101D1 | | Descriptor: | CAMPHOR, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
2FDP
 
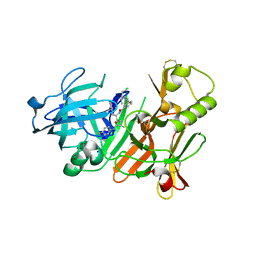 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
3LXH
 
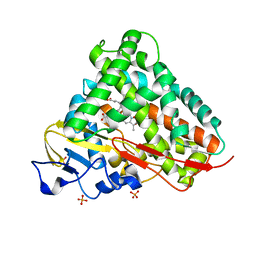 | | Crystal Structure of Cytochrome P450 CYP101D1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3LXD
 
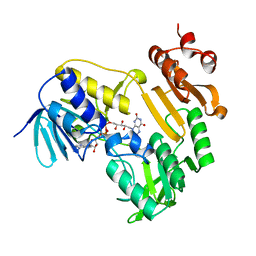 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | Descriptor: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
3NV6
 
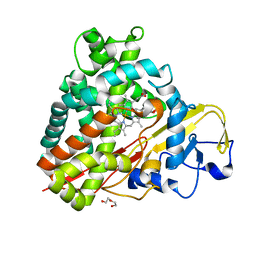 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3NV5
 
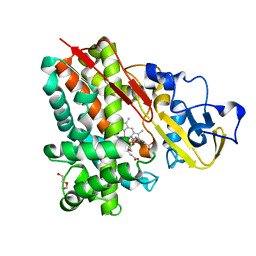 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3SYY
 
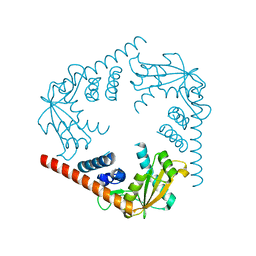 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ4
 
 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ5
 
 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
6B3X
 
 | | Crystal structure of CstF-50 in complex with CstF-77 | | Descriptor: | Cleavage stimulation factor subunit 1, Cleavage stimulation factor subunit 3 | | Authors: | Yang, W, Hsu, P, Yang, F, Song, J.E, Varani, G. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Nucleic Acids Res., 46, 2018
|
|
7WWH
 
 | |
1B63
 
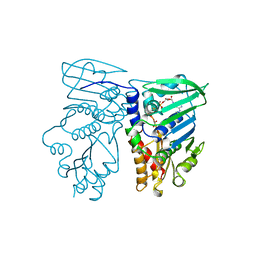 | | MUTL COMPLEXED WITH ADPNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MUTL, ... | | Authors: | Yang, W. | | Deposit date: | 1999-01-20 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair.
Cell(Cambridge,Mass.), 97, 1999
|
|
1AZO
 
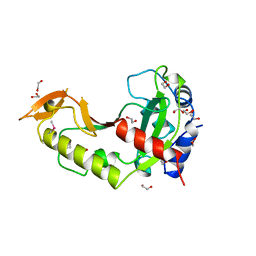 | | DNA MISMATCH REPAIR PROTEIN MUTH FROM E. COLI | | Descriptor: | 1,2-ETHANEDIOL, MUTH | | Authors: | Yang, W. | | Deposit date: | 1997-11-19 | | Release date: | 1998-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases.
EMBO J., 17, 1998
|
|
1BKN
 
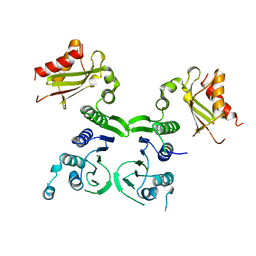 | |
2AZO
 
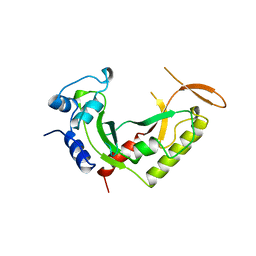 | | DNA MISMATCH REPAIR PROTEIN MUTH FROM E. COLI | | Descriptor: | MUTH | | Authors: | Yang, W. | | Deposit date: | 1997-11-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases.
EMBO J., 17, 1998
|
|
3PSX
 
 | | Crystal structure of the KT2 mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Yorke, J.A, Bell, S.G, Zhou, W, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure, electronic properties and catalytic behaviour of an activity-enhancing CYP102A1 (P450(BM3)) variant
Dalton Trans, 40, 2011
|
|
3TKT
 
 | | Crystal structure of CYP108D1 from Novosphingobium aromaticivorans DSM12444 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2011-08-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of CYP108D1 from Novosphingobium aromaticivorans DSM12444: an aromatic hydrocarbon-binding P450 enzyme
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HF2
 
 | | Crystal structure of the I401P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2009-05-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Highly Active Single-Mutation Variant of P450(BM3) (CYP102A1)
Chembiochem, 10, 2009
|
|
3K5O
 
 | | Crystal structure of E.coli Pol II | | Descriptor: | DNA polymerase II | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
