8P5W
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5S
 
 | | Crystal structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Boyko, A, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5V
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5U
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5X
 
 | | Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, MAGNESIUM ION, Oxoglutarate dehydrogenase inhibitor, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
2WFL
 
 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WFM
 
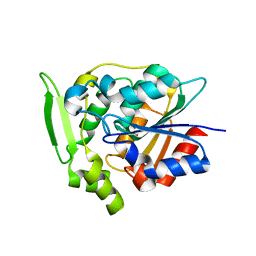 | | Crystal structure of polyneuridine aldehyde esterase mutant (H244A) | | Descriptor: | POLYNEURIDINE ALDEHYDE ESTERASE | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
8P1U
 
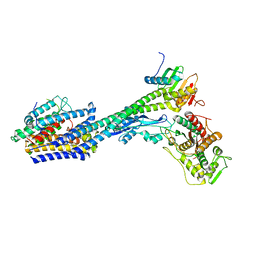 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
4C1X
 
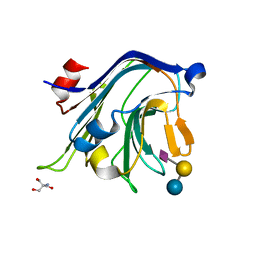 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 6'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, SIALIDASE A | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Characterization of the Carbohydrate-Binding Module of Nana Sialidase, a Pneumococcal Virulence Factor.
Bmc Struct.Biol., 15, 2015
|
|
4ZXK
 
 | |
4K9R
 
 | | Spore photoproduct lyase Y98F mutant | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Spore photoproduct lyase, ... | | Authors: | Yang, L, Nelson, R.S, Benjdia, A, Lin, G, Telser, J, Stoll, S, Schlichting, I, Li, L. | | Deposit date: | 2013-04-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A radical transfer pathway in spore photoproduct lyase.
Biochemistry, 52, 2013
|
|
2LS4
 
 | |
2LS2
 
 | |
2LS3
 
 | |
7YPN
 
 | | Crystal structure of transaminase CC1012 mutant M9 complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7YPM
 
 | | Crystal structure of transaminase CC1012 complexed with PLP and L-alanine | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aspartate aminotransferase family protein, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8XXC
 
 | | DaCS-intermediate complex | | Descriptor: | ACETIC ACID, CITRIC ACID, Citrate synthase, ... | | Authors: | Yang, L, Fang, Y.J. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | DaCS-intermediate complex
To Be Published
|
|
8XXD
 
 | | TtCS-citrate-CoA complex | | Descriptor: | CITRIC ACID, COENZYME A, Citrate synthase, ... | | Authors: | Yang, L, Fang, Y.J. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | TtCS-citrate-CoA complex
To Be Published
|
|
8XXF
 
 | | TtCS, oxaloacetate, acetyl-CoA complex | | Descriptor: | ACETYL COENZYME *A, COENZYME A, Citrate synthase, ... | | Authors: | Yang, L, Fang, Y.J. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | TtCS, oxaloacetate, acetyl-CoA complex
To Be Published
|
|
8XU1
 
 | | DaCS-oxaloacetate complex | | Descriptor: | OXALOACETATE ION, citrate synthase | | Authors: | Yang, L, Fang, Y.J. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | DaCS-oxaloacetate complex
To Be Published
|
|
8XXE
 
 | | TtCS-intermediate complex | | Descriptor: | ACETYL GROUP, COENZYME A, Citrate synthase, ... | | Authors: | Yang, L, Fang, Y.J. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | TtCS-intermediate complex
To Be Published
|
|
7WBR
 
 | |
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5WWX
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(P*AP*GP*AP*GP*U)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
