9J6I
 
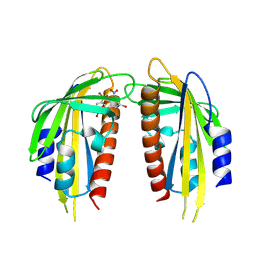 | | Crystal structure of the ABA receptor PYL1 in complex with DBSA compound | | Descriptor: | 5-bromanyl-N2-[(4-bromophenyl)-bis(oxidanyl)-$l^4-sulfanyl]-N1,N1,N3,N3-tetrakis(oxidanyl)benzene-1,2,3-triamine, Abscisic acid receptor PYL1 | | Authors: | Yan, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemical control of plant water loss by stabilizing dimeric ABA receptor
To Be Published
|
|
2ADZ
 
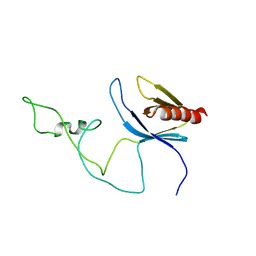 | | solution structure of the joined PH domain of alpha1-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Wen, W, Xu, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
5N41
 
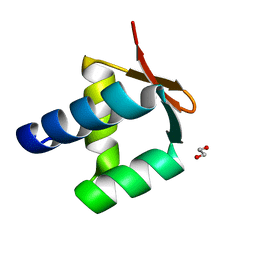 | | Archaeal DNA polymerase holoenzyme - SSO6202 at 1.35 Ang resolution | | Descriptor: | 1,2-ETHANEDIOL, PolB1 Binding Protein 2 | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
5N35
 
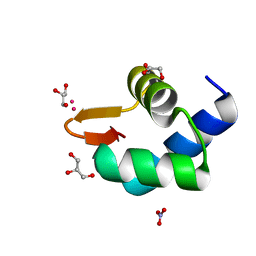 | | Gadolinium phased PBP2 (SSO6202) at 2.2 Ang | | Descriptor: | GADOLINIUM ATOM, GLYCEROL, NITRATE ION, ... | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-17 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
8JXZ
 
 | | Chitin binding SusD-like protein AqSusD in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SusD-like protein AqSusD | | Authors: | Yang, J. | | Deposit date: | 2023-07-01 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
5IZW
 
 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
8J0P
 
 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
2D2C
 
 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1Z87
 
 | | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1Z86
 
 | | Solution structure of the PDZ domain of alpha-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
5GI0
 
 | | Crystal structure of RNA editing factor MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5IWW
 
 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
5IWB
 
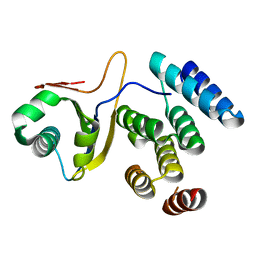 | | Crystal structure of design pentatricopeptide repeat complex with MORF protein | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS3-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis for designer pentatricopeptide repeat proteins interaction with MORF protein
To be published
|
|
1H6G
 
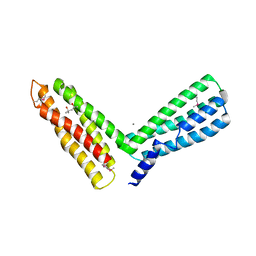 | | alpha-catenin M-domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-1 CATENIN, CALCIUM ION, ... | | Authors: | Yang, J, Dokurno, P, Tonks, N.K, Barford, D. | | Deposit date: | 2001-06-14 | | Release date: | 2001-08-07 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the M-Fragment of Alpha-Catenin: Implications for Modulation of Cell Adhesion.
Embo J., 20, 2001
|
|
3RVZ
 
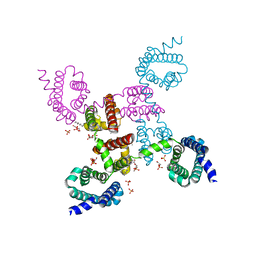 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RW0
 
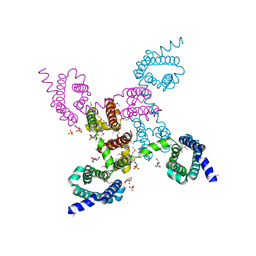 | | Crystal structure of the NavAb voltage-gated sodium channel (Met221Cys, 2.95 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
1GWZ
 
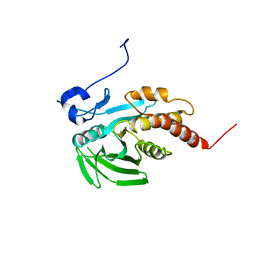 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
2KNC
 
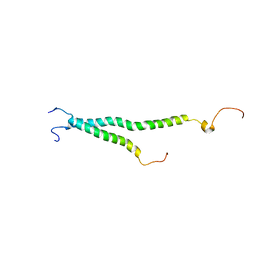 | | Platelet integrin ALFAIIB-BETA3 transmembrane-cytoplasmic heterocomplex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Yang, J, Ma, Y, Page, R.C, Misra, S, Plow, E.F, Qin, J. | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ASH
 
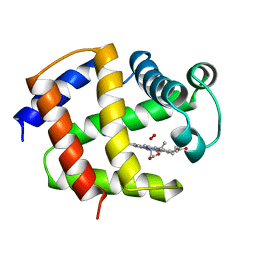 | | THE STRUCTURE OF ASCARIS HEMOGLOBIN DOMAIN I AT 2.2 ANGSTROMS RESOLUTION: MOLECULAR FEATURES OF OXYGEN AVIDITY | | Descriptor: | HEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, J, Mathews, F.S, Kloek, A.P, Goldberg, D.E. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7YP0
 
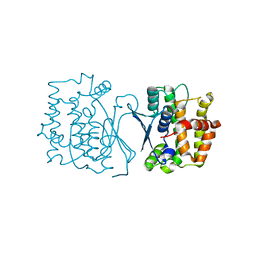 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
2LR7
 
 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
2TPR
 
 | |
6PZP
 
 | | Crystal structure of caspase-1 in complex with VX-765 | | Descriptor: | Caspase-1, N-(4-amino-3-chlorobenzene-1-carbonyl)-3-methyl-L-valyl-N-[(2S)-1-carboxy-3-oxopropan-2-yl]-L-prolinamide | | Authors: | Yang, J, Liu, Z, Xiao, T.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of caspase-1 in complex with VX-765
To Be Published
|
|
8K2P
 
 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
