251L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
247L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
237L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
241L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
249L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
236L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
244L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
248L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
239L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
243L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
250L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5XNB
 
 | |
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1C1E
 
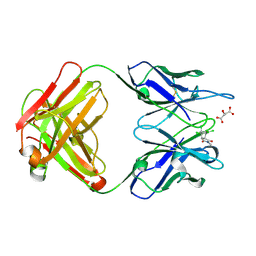 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
7ADZ
 
 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state. | | Descriptor: | Phage tail protein, Putative phage tail sheath protein FI, cap adaptor protein (Algo2), ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
7AEB
 
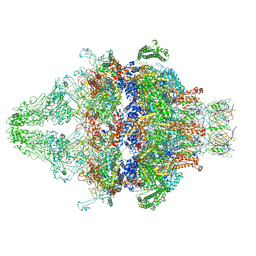 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry. | | Descriptor: | Baseplate_J domain-containing protein, LysM domain-containing protein, Phage tail protein, ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
7AE0
 
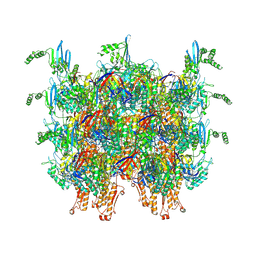 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the sheath-tube module in extended state. | | Descriptor: | Phage tail protein, Putative phage tail sheath protein FI | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
7AEF
 
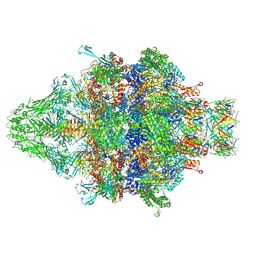 | | Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry. | | Descriptor: | Baseplate_J domain-containing protein, LysM domain-containing protein, Phage tail protein, ... | | Authors: | Xu, J, Ericson, C, Feldmueller, M, Lien, Y.W, Pilhofer, M. | | Deposit date: | 2020-09-17 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Identification and structure of an extracellular contractile injection system from the marine bacterium Algoriphagus machipongonensis.
Nat Microbiol, 7, 2022
|
|
3WDH
 
 | | Crystal structure of Pullulanase from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WDI
 
 | | Crystal structure of Pullulanase complexed with maltotriose from Anoxybacillus sp. LM18-11 | | Descriptor: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
4IKD
 
 | | Crystal structure of SNX11 PX domain | | Descriptor: | CHLORIDE ION, SODIUM ION, Sorting nexin-11 | | Authors: | Xu, J, Xu, T, Liu, J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Sorting Nexin 11 (SNX11) Reveals a Novel Extended PX Domain (PXe Domain) Critical for the Inhibition of Sorting Nexin 10 (SNX10) Induced Vacuolation
to be published
|
|
4IKB
 
 | | Crystal structure of SNX11 PX domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Xu, J, Xu, T, Liu, J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Sorting Nexin 11 (SNX11) Reveals a Novel Extended PX Domain (PXe Domain) Critical for the Inhibition of Sorting Nexin 10 (SNX10) Induced Vacuolation
to be published
|
|
6QX7
 
 | |
6QZ9
 
 | | The cryo-EM structure of the collar complex and tail axis in bacteriophage phi29 | | Descriptor: | Portal protein, Pre-neck appendage protein, Proximal tail tube connector protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
6QYZ
 
 | |
