6HWH
 
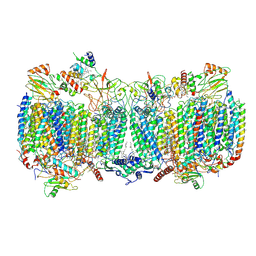 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | Descriptor: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | Authors: | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-11-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4JFB
 
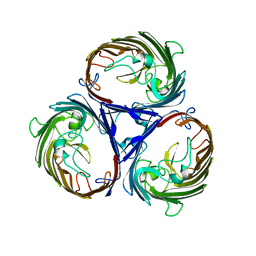 | | Crystal structure of OmpF in C2 with tNCS | | Descriptor: | Outer membrane protein F | | Authors: | Wiseman, B, Kilburg, A, Chaptal, V, Reyes-Meija, G.C, Sarwan, J, Falson, P, Jault, J.M. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Stubborn contaminants: influence of detergents on the purity of the multidrug ABC transporter BmrA
PLoS ONE, 9, 2014
|
|
8P3O
 
 | |
8P3P
 
 | |
6NWR
 
 | |
8BHW
 
 | |
6Q3A
 
 | | Apo form of Apolipoprotein N-acyltransferase (Lnt) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Wiseman, B, Hogbom, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes in Apolipoprotein N-acyltransferase (Lnt).
Sci Rep, 10, 2020
|
|
6RBG
 
 | |
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
7BG4
 
 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP, Mg, and Rhodamine 6G solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6R81
 
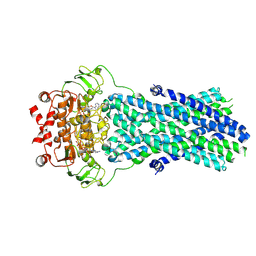 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
4D5U
 
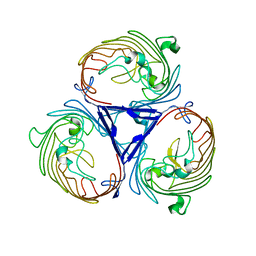 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
5SXR
 
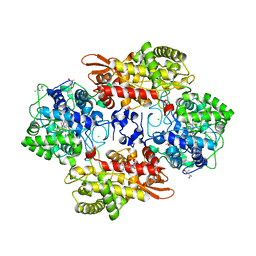 | | Crystal structure of B. pseudomallei KatG with NAD bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases.
J. Biol. Chem., 285, 2010
|
|
5SXQ
 
 | |
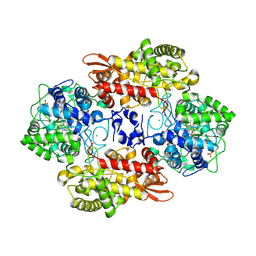
5SXX
 
 | |
5SXS
 
 | |
5SXW
 
 | |
5SXT
 
 | |
8RKE
 
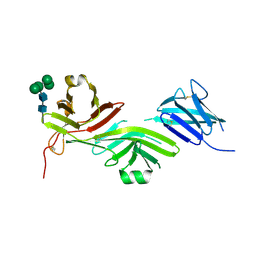 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKF
 
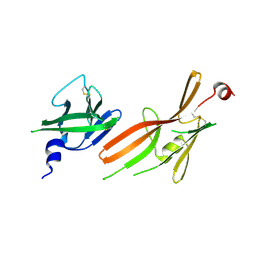 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKH
 
 | | Crystal structure of the ZP-N2 and ZP-N3 domains of mouse ZP2 (mZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKG
 
 | | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICINE, ... | | Authors: | Nishio, S, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8BQU
 
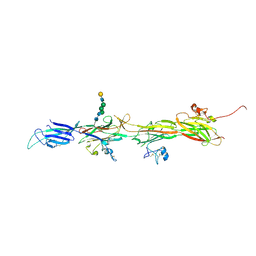 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
5L02
 
 | | S324T variant of B. pseudomallei KatG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
