8HUH
 
 | | Crystal structure of T2R-TTL-3a complex | | Descriptor: | 2-(1-methylindol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1~{H}-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y.X, Chen, J.J. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of T2R-TTL-3a complex
To Be Published
|
|
6LLK
 
 | | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
6LL0
 
 | | Carbazole-soaked reduced oxygenase in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 9H-CARBAZOLE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbazole-soaked reduced oxygenase in carbazole 1,9a-dioxygenase
To Be Published
|
|
6LL4
 
 | | Oxygen-exposed carbazole-soaked reduced terminal oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | (9aR)-9a-(dioxidanyl)-1,9-dihydrocarbazole, 1,2-ETHANEDIOL, 2'-amino[1,1'-biphenyl]-2,3-diol, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-exposed carbazole-soaked reduced terminal oxygenase of carbazole 1,9a-dioxygenase with
To Be Published
|
|
6LLM
 
 | | Biphenyl-2,3-diol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BIPHENYL-2,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biphenyl-2,3-diol-soaked terminal oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
6LL1
 
 | | Oxygen-exposed reduced terminal oxygenase in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxygen-exposed reduced terminal oxygenase in carbazole 1,9a-dioxygenase
To Be Published
|
|
6LLH
 
 | | Biphenyl-2,3-diol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BIPHENYL-2,3-DIOL, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase
To Be Published
|
|
8HY7
 
 | | EGFR kinase domain mutant "TMLR" with compound 28f | | Descriptor: | Epidermal growth factor receptor, ~{N}-[3-[[6-fluoranyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]quinazolin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The crystal structure of EGFR kinase domain(T790M/L858R) in complex with compound 28f
To Be Published
|
|
6LLF
 
 | | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2019-12-23 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase
To Be Published
|
|
8ZOW
 
 | | Cryo-EM structure of Metyltetraprole-bound porcine bc1 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1-[2-[[1-(4-chlorophenyl)pyrazol-3-yl]oxymethyl]-3-methyl-phenyl]-4-methyl-1,2,3,4-tetrazol-5-one, CARDIOLIPIN, ... | | Authors: | Wang, Y.X, Sun, J.Y, Cui, G.R, Yang, G.F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8ZOS
 
 | | Cryo-EM structure of pyraclostrobin-bound porcine bc1 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Complex III subunit 9, ... | | Authors: | Wang, Y.X, Sun, J.Y, Li, Z.W, Cui, G.R, Yang, G.F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8ZP0
 
 | | Cryo-EM structure of YF23694-bound porcine bc1 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1-[2-[(4,6-dimethyl-1,3-benzothiazol-2-yl)sulfanylmethyl]-3-methyl-phenyl]-4-methyl-1,2,3,4-tetrazol-5-one, CARDIOLIPIN, ... | | Authors: | Wang, Y.X, Sun, J.Y, Cui, G.R, Yang, G.F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8IOG
 
 | |
7DMZ
 
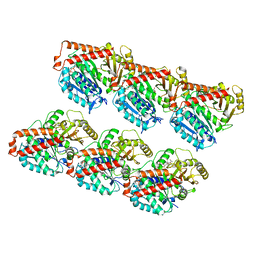 | | GMPCPP microtubule complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Wang, Y.X. | | Deposit date: | 2020-12-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | GMPCPP microtubule complex
To Be Published
|
|
7DN0
 
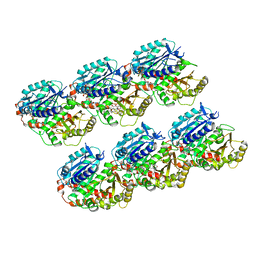 | | AJ-GMPCPP-MT-non-seam | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Wang, Y.X. | | Deposit date: | 2020-12-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | GMPCPP microtubule
To Be Published
|
|
5JQG
 
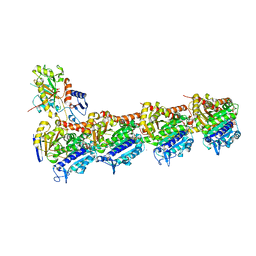 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Naismith, J.H, Zhu, X. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
9LNW
 
 | |
9LNV
 
 | |
9LNX
 
 | |
9LNL
 
 | |
8ZOM
 
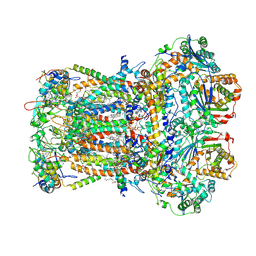 | | Cryo-EM structure of pyraclostrobin-bound Arachis hypogaea bc1 complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Cui, G.R, Wang, Y.X, Yang, G.F. | | Deposit date: | 2024-05-28 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structure of pyraclostrobin-bound Arachis hypogaea bc1 complex
To Be Published
|
|
1S2F
 
 | | Average solution structure of a pseudo-5'-splice site from the negative regulator of splicing of Rous Sarcoma virus | | Descriptor: | 5'-R(*GP*GP*GP*GP*AP*GP*UP*GP*GP*UP*UP*UP*GP*UP*AP*UP*CP*CP*UP*UP*CP*CP*C)-3' | | Authors: | Cabello-Villegas, J, Giles, K.E, Soto, A.M, Yu, P, Beemon, K.L, Wang, Y.X. | | Deposit date: | 2004-01-08 | | Release date: | 2004-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor.
Rna, 10, 2004
|
|
1S34
 
 | | Solution structure of residues 907-929 from Rous Sarcoma Virus | | Descriptor: | 5'-R(*GP*GP*GP*GP*AP*GP*UP*GP*GP*UP*UP*UP*GP*UP*AP*UP*CP*CP*UP*UP*CP*CP*C)-3' | | Authors: | Cabello-Villegas, J, Giles, K.E, Soto, A.M, Yu, P, Beemon, K.L, Wang, Y.X. | | Deposit date: | 2004-01-12 | | Release date: | 2004-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor.
Rna, 10, 2004
|
|
1OV2
 
 | | Ensemble of the solution structures of domain one of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
2E34
 
 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
