4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZO0
 
 | | Neutron structure of xylanase at pD5.4 | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Wan, Q, Li, Z.H. | | Deposit date: | 2018-04-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.648 Å) | | Cite: | Neutron structure of xylanase at pD5.4
To be published
|
|
3NVO
 
 | |
3NWI
 
 | |
4PDJ
 
 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7D49
 
 | |
7D3Z
 
 | |
7D4L
 
 | |
7D4X
 
 | |
7D6G
 
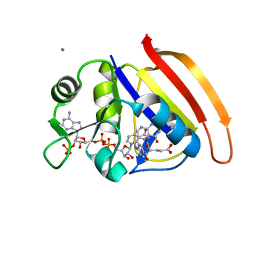 | |
4HKL
 
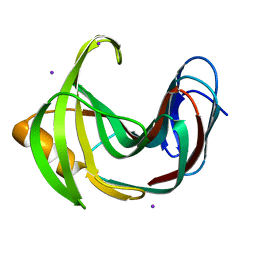 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKO
 
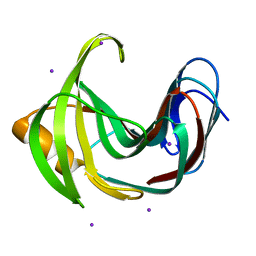 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKW
 
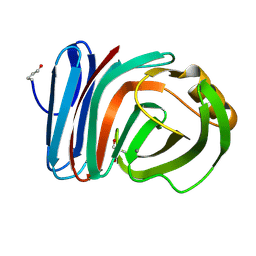 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with Substrate and Products | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endo-1,4-beta-xylanase 2, ... | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P, Coates, L. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK8
 
 | | Crystal Structures of Mutant Endo- -1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | CITRIC ACID, Endo-1,4-beta-xylanase 2, GLYCEROL, ... | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK9
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RGC
 
 | | 277K Crystal structure of Escherichia Coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennett, B.C, Dealwis, C. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4S2G
 
 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 5.8 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Kovalevsky, A, Wan, Q, Langan, P. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-25 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZKZ
 
 | |
5ZIW
 
 | |
4S2D
 
 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II in complex with MES at pH 5.7 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-25 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4S2F
 
 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II at pH 4.4 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Kovalevsky, A, Wan, Q, Langan, P. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2019-12-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5ZII
 
 | |
