3RHQ
 
 | |
3RHM
 
 | |
3RHJ
 
 | |
3RHL
 
 | |
3RHR
 
 | |
7RLU
 
 | | Structure of ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) in complex with NADP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
7RLT
 
 | | Structure of ligand-free ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
4GO0
 
 | |
4GO2
 
 | |
4GNZ
 
 | |
3RHO
 
 | |
3RHP
 
 | |
2O2Q
 
 | | Crystal structure of the C-terminal domain of rat 10'formyltetrahydrofolate dehydrogenase in complex with NADP | | Descriptor: | Formyltetrahydrofolate dehydrogenase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tsybovsky, Y, Donato, H, Krupenko, N.I, Davies, C, Krupenko, S.A. | | Deposit date: | 2006-11-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases.
Biochemistry, 46, 2007
|
|
2O2P
 
 | | Crystal structure of the C-terminal domain of rat 10'formyltetrahydrofolate dehydrogenase | | Descriptor: | Formyltetrahydrofolate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Tsybovsky, Y, Donato, H, Krupenko, N.I, Davies, C, Krupenko, S.A. | | Deposit date: | 2006-11-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases.
Biochemistry, 46, 2007
|
|
2O2R
 
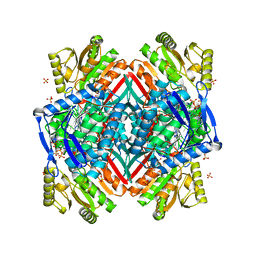 | | Crystal structure of the C-terminal domain of rat 10'formyltetrahydrofolate dehydrogenase in complex with NADPH | | Descriptor: | Formyltetrahydrofolate dehydrogenase, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tsybovsky, Y, Donato, H, Krupenko, N.I, Davies, C, Krupenko, S.A. | | Deposit date: | 2006-11-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases.
Biochemistry, 46, 2007
|
|
8GAT
 
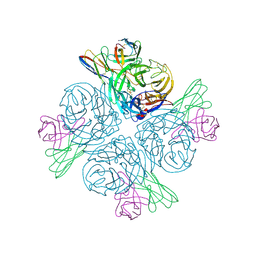 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAU
 
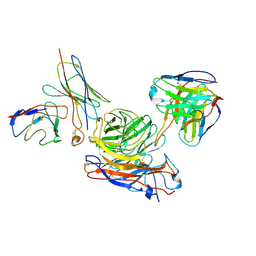 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAV
 
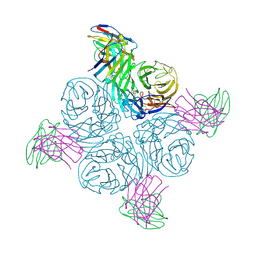 | | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab NDS.3, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8EEU
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05 | | Descriptor: | Coat protein, Fab SKT05 heavy chain, Fab SKT05 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
7MTD
 
 | | Structure of aged SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MTC
 
 | | Structure of freshly purified SARS-CoV-2 S2P spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tsybovsky, Y, Olia, A.S, Kwong, P.D. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity.
J.Biol.Chem., 297, 2021
|
|
7MTE
 
 | |
7U9P
 
 | | SARS-CoV-2 spike trimer RBD in complex with Fab NA8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NA8 Fab heavy chain, NA8 Fab light chain, ... | | Authors: | Tsybovsky, Y, Kwong, P.D, Farci, P. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants.
Cell Rep, 41, 2022
|
|
7U9O
 
 | | SARS-CoV-2 spike trimer RBD in complex with Fab NE12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NE12 Fab heavy chain, NE12 Fab light chain, ... | | Authors: | Tsybovsky, Y, Kwong, P.D, Farci, P. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants.
Cell Rep, 41, 2022
|
|
