4UQT
 
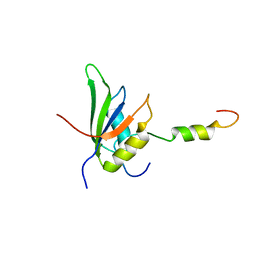 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
2JPD
 
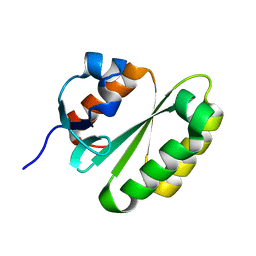 | | Solution structure of the ERCC1 central domain | | Descriptor: | DNA excision repair protein ERCC-1 | | Authors: | Tripsianes, K, Folkers, G, Zheng, C, Das, D, Grinstead, J.S, Kaptein, R, Boelens, R. | | Deposit date: | 2007-05-06 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization
Nucleic Acids Res., 35, 2007
|
|
1Z00
 
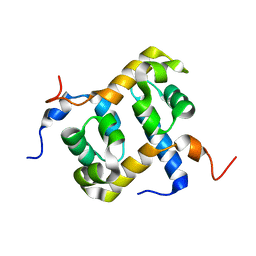 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
4A4F
 
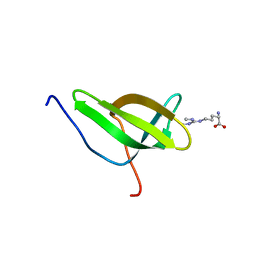 | | Solution structure of SPF30 Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4E
 
 | | Solution structure of SMN Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL MOTOR NEURON PROTEIN | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4H
 
 | | Solution structure of SPF30 Tudor domain in complex with asymmetrically dimethylated arginine | | Descriptor: | NG,NG-DIMETHYL-L-ARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4G
 
 | | Solution structure of SMN Tudor domain in complex with asymmetrically dimethylated arginine | | Descriptor: | NG,NG-DIMETHYL-L-ARGININE, SURVIVAL MOTOR NEURON PROTEIN | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
5MMC
 
 | |
5WOT
 
 | |
5WOY
 
 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4EL6
 
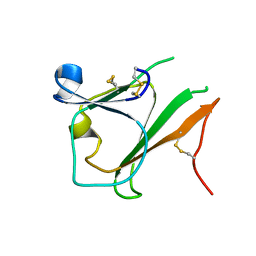 | | Crystal structure of IPSE/alpha-1 from Schistosoma mansoni eggs | | Descriptor: | IL-4-inducing protein | | Authors: | Mayerhofer, H, Meyer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Sattler, M, Schramm, G, Mueller-Dieckmann, J. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and functional analysis of IPSE/alpha-1, an IL-4-inducing factor secreted from Schistosoma mansoni eggs, reveals an IgE-binding crystallin fold
To be Published
|
|
6YEU
 
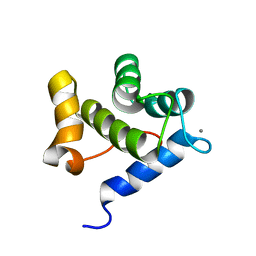 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YET
 
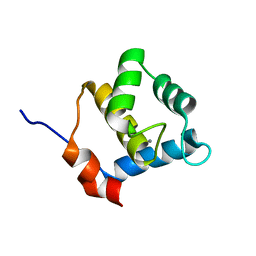 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
2AQ0
 
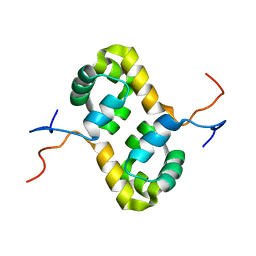 | | Solution structure of the human homodimeric dna repair protein XPF | | Descriptor: | DNA repair endonuclease XPF | | Authors: | Das, D, Tripsianes, K, Folkers, G, Jaspers, N.G, Hoeijmakers, J.H, Kaptein, R, Boelens, R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The HhH domain of the human DNA repair protein XPF forms stable homodimers
Proteins, 70, 2008
|
|
2XA6
 
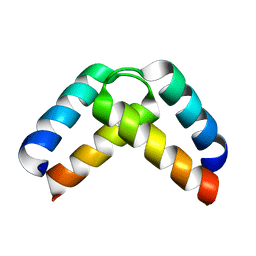 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
5AGQ
 
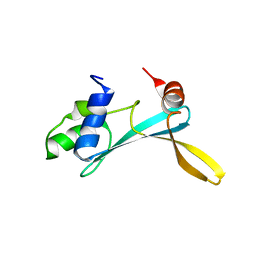 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5WOX
 
 | |
5WOZ
 
 | |
5MGQ
 
 | |
7OX6
 
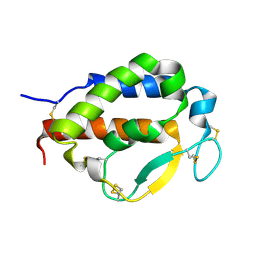 | | Solution structure of human interleukin-9 | | Descriptor: | Interleukin-9 | | Authors: | Savvides, S.N, Tripsianes, K, De Vos, T, Papageorgiou, A, Evangelidis, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-11 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
5LNF
 
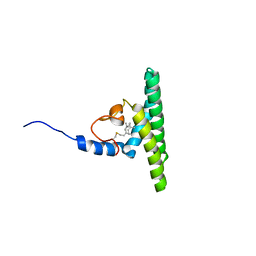 | | Solution NMR structure of farnesylated PEX19, C-terminal domain | | Descriptor: | FARNESYL, Peroxisomal biogenesis factor 19 | | Authors: | Emmanouilidis, L, Schuetz, U, Tripsianes, K, Madl, T, Radke, J, Rucktaeschel, R, Wilmanns, M, Schliebs, W, Erdmann, R, Sattler, M. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-15 | | Last modified: | 2019-09-11 | | Method: | SOLUTION NMR | | Cite: | Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19.
Nat Commun, 8, 2017
|
|
4B9W
 
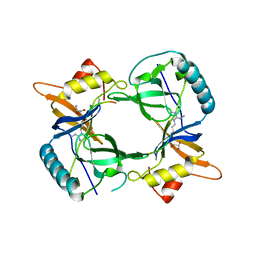 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | Descriptor: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
4A24
 
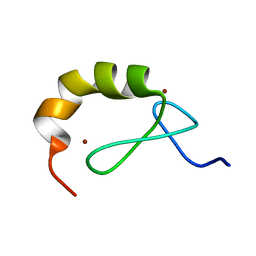 | | Structural and functional analysis of the DEAF-1 and BS69 MYND domains | | Descriptor: | DEFORMED EPIDERMAL AUTOREGULATORY FACTOR 1 HOMOLOG, ZINC ION | | Authors: | Kateb, F, Perrin, H, Tripsianes, K, Zou, P, Spadaccini, R, Bottomley, M, Bepperling, A, Ansieau, S, Sattler, M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Plos One, 8, 2013
|
|
4B9X
 
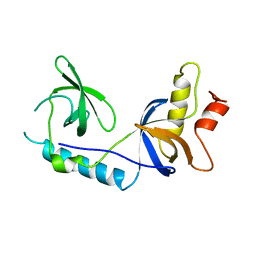 | | Structure of extended Tudor domain TD3 from mouse TDRD1 | | Descriptor: | TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Multiple Tudor Domain-Containing Protein Tdrd1 is a Molecular Scaffold for Mouse Piwi Proteins and Pirna Biogenesis Factors.
RNA, 18, 2012
|
|
