7U0L
 
 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US9
 
 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USB
 
 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US6
 
 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USA
 
 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7K45
 
 | |
4CBL
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBH
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBM
 
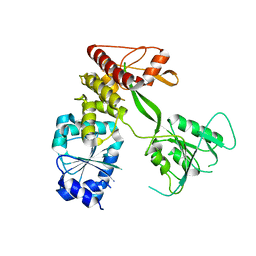 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBI
 
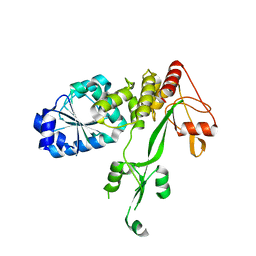 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBG
 
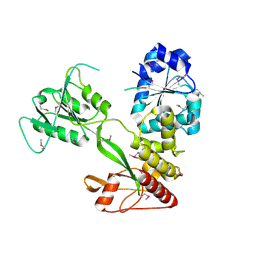 | | Pestivirus NS3 helicase | | Descriptor: | ACETATE ION, SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
7K43
 
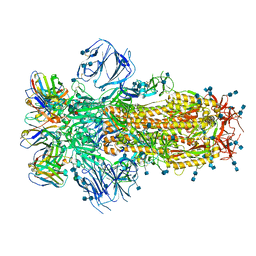 | |
9AU2
 
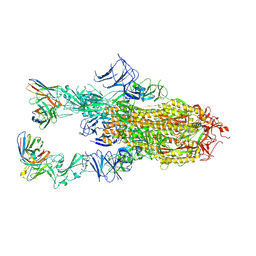 | | VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Park, Y.J, Veelser, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
7U6R
 
 | |
7U0A
 
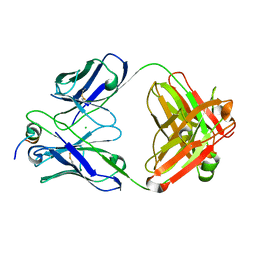 | |
7U09
 
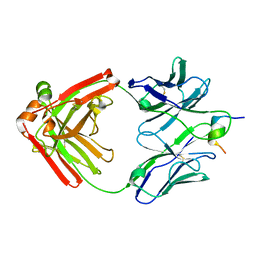 | |
7U0E
 
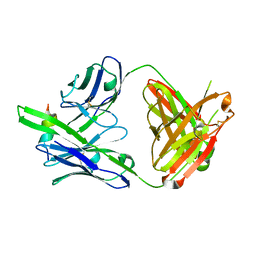 | |
7RA8
 
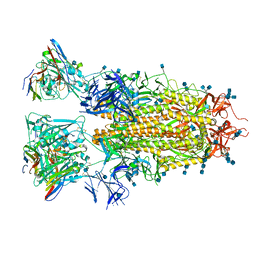 | |
7RAL
 
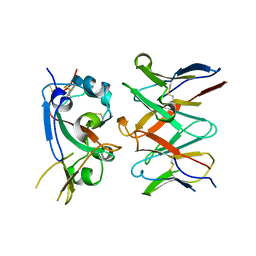 | |
5MZ4
 
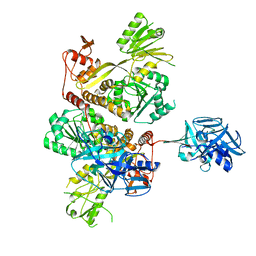 | | Crystal Structure of full-lengh CSFV NS3/4A | | Descriptor: | Genome polyprotein,Genome polyprotein | | Authors: | Tortorici, M.A, Rey, F.A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.048 Å) | | Cite: | A positive-strand RNA virus uses alternative protein-protein interactions within a viral protease/cofactor complex to switch between RNA replication and virion morphogenesis.
PLoS Pathog., 13, 2017
|
|
7SL5
 
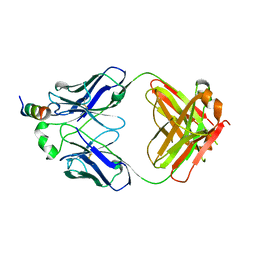 | |
7SKZ
 
 | |
9DGO
 
 | |
7JXE
 
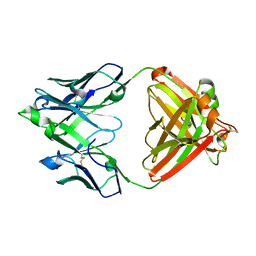 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | S2X35 antigen-binding (Fab) fragment | | Authors: | Tortorici, M.A, Park, Y.J, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
9ASD
 
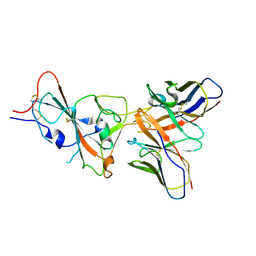 | | VIR-7229 Fab fragment bound the SARS-CoV-2 BA.2.86 spike trimer (local refinement of the BA 2.86 RBD/VIR-7229 VHVL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VIR-7229 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
