8BA1
 
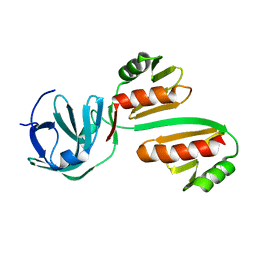 | | CTD12-CTD12 heterodimer from CPSF73 and CPSF100 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
8B7T
 
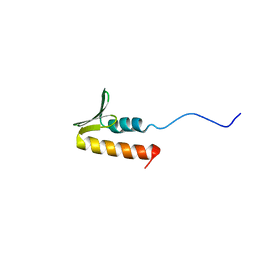 | | CPSF73 CTD3 | | Descriptor: | CPSF73 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
2CKY
 
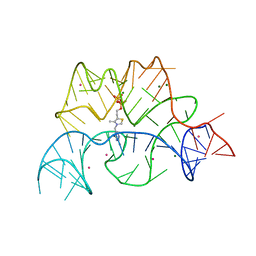 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
1M8V
 
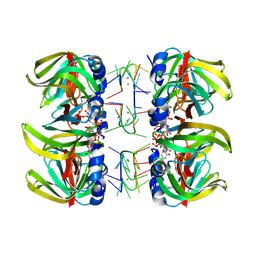 | | Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', CALCIUM ION, PUTATIVE SNRNP SM-LIKE PROTEIN, ... | | Authors: | Thore, S, Mayer, C, Sauter, C, Weeks, S, Suck, D. | | Deposit date: | 2002-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pyrococcus abyssii Sm core and its Complex with RNA: Common Features of RNA-binding in Archaea and Eukarya
J.Biol.Chem., 278, 2003
|
|
1UOC
 
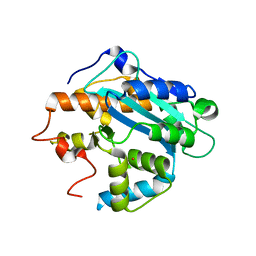 | | X-ray structure of the RNase domain of the yeast Pop2 protein | | Descriptor: | CALCIUM ION, POP2, XENON | | Authors: | Thore, S, Mauxion, F, Seraphin, B, Suck, D. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure and Activity of the Yeast Pop2 Protein: A Nuclease Subunit of the Mrna Deadenylase Complex
Embo Rep., 4, 2003
|
|
3D2V
 
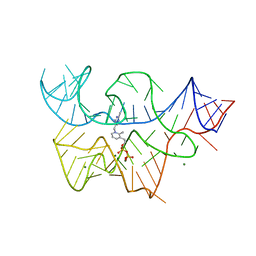 | |
3D2G
 
 | |
3D2X
 
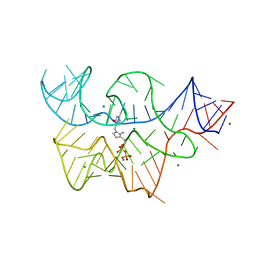 | | Structure of the thiamine pyrophosphate-specific riboswitch bound to oxythiamine pyrophosphate | | Descriptor: | 3-[(4-hydroxy-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, MAGNESIUM ION, TPP-specific riboswitch | | Authors: | Thore, S, Frick, C, Ban, N. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch
J.Am.Chem.Soc., 130, 2008
|
|
5NLG
 
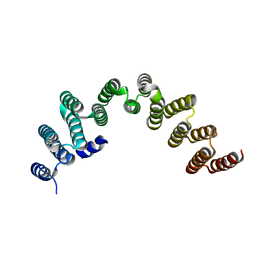 | | RRP5 C-terminal domain | | Descriptor: | rRNA biogenesis protein RRP5 | | Authors: | Thore, S, Fribourg, S. | | Deposit date: | 2017-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and interaction analysis of the Rrp5 C-terminal region.
FEBS Open Bio, 8, 2018
|
|
2JJ6
 
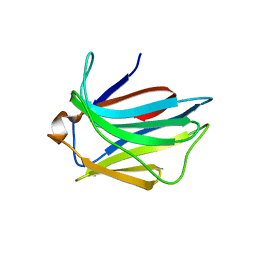 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
5HZD
 
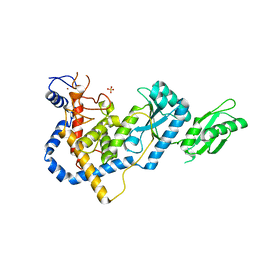 | | RNA Editing TUTase 1 from Trypanosoma brucei | | Descriptor: | 3' terminal uridylyl transferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5I49
 
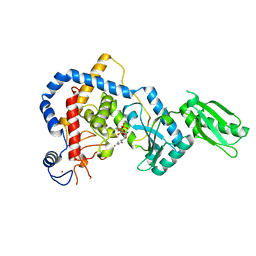 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP analog UMPNPP | | Descriptor: | 3' terminal uridylyl transferase, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5IDO
 
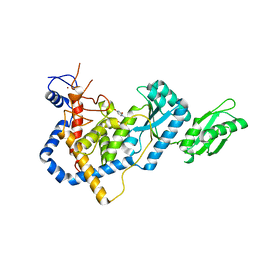 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
4WN4
 
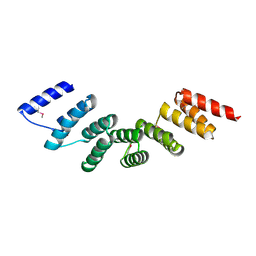 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WSL
 
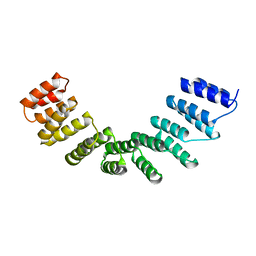 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
3VEM
 
 | | Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1 | | Descriptor: | Helicase protein MOM1 | | Authors: | Nishikura, T, Petty, T.J, Halazonetis, T, Paszkowski, J, Thore, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Transcriptional Gene Silencing Mediated by Arabidopsis MOM1.
PLOS GENET., 8, 2012
|
|
4R7N
 
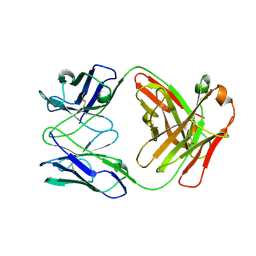 | | Fab C2E3 | | Descriptor: | Fab C2E3 Heavy chain, Fab C2E3 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-28 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
4R7D
 
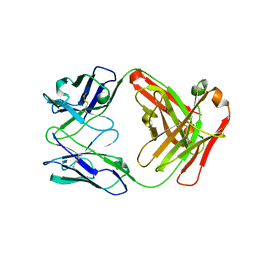 | | Fab Hu 15C1 | | Descriptor: | Fab Hu 15C1 Heavy chain, Fab Hu 15C1 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
4H67
 
 | | Crystal structure of HMP synthase Thi5 from S. cerevisiae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyrimidine precursor biosynthesis enzyme THI5, SULFATE ION | | Authors: | Coquille, S.C, Roux, C, Fitzpatrick, T, Thore, S. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Last Piece in the Vitamin B1 Biosynthesis Puzzle: STRUCTURAL AND FUNCTIONAL INSIGHT INTO YEAST 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE PHOSPHATE (HMP-P) SYNTHASE.
J.Biol.Chem., 287, 2012
|
|
4H65
 
 | |
4H6D
 
 | | Crystal structure of PLP-soaked HMP synthase Thi5 from S. cerevisiae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyrimidine precursor biosynthesis enzyme THI5 | | Authors: | Coquille, S.C, Roux, C, Fitzpatrick, T, Thore, S. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Last Piece in the Vitamin B1 Biosynthesis Puzzle: STRUCTURAL AND FUNCTIONAL INSIGHT INTO YEAST 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE PHOSPHATE (HMP-P) SYNTHASE.
J.Biol.Chem., 287, 2012
|
|
6FTU
 
 | | Structure of a Quadruplex forming sequence from D. discoideum | | Descriptor: | DNA (26-MER), POTASSIUM ION | | Authors: | Guedin, A, Linda, L, Armane, S, Lacroix, L, Mergny, J.L, Thore, S, Yatsunyk, L.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Quadruplexes in 'Dicty': crystal structure of a four-quartet G-quadruplex formed by G-rich motif found in the Dictyostelium discoideum genome.
Nucleic Acids Res., 46, 2018
|
|
6HYE
 
 | | PDX1.2/PDX1.3 complex (PDX1.3:K97A) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HXG
 
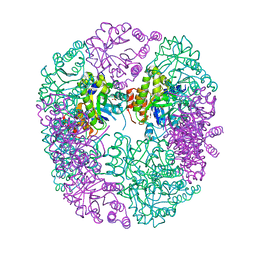 | | PDX1.2/PDX1.3 complex (intermediate) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6HX3
 
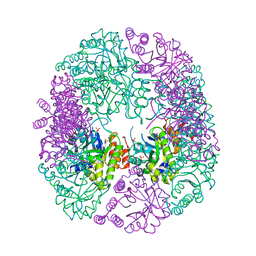 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
