9J6K
 
 | | HBx complexed with DDB1 | | Descriptor: | DNA damage-binding protein 1, Protein X | | Authors: | Tanaka, H, Kita, S, Sasaki, M, Maenaka, K, Machida, S. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of the hepatitis B virus X protein in complex with DDB1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
2RSY
 
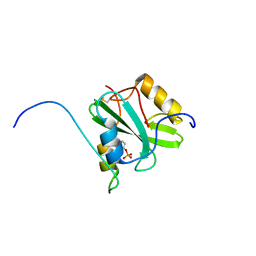 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
6KVD
 
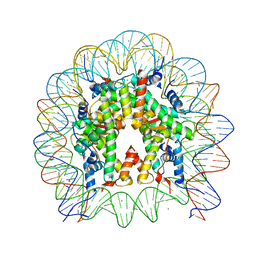 | | Crystal structure of human nucleosome containing H2A.J | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.J, ... | | Authors: | Tanaka, H, Koyama, M, Sato, S, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and structural analyses of the nucleosome containing human histone H2A.J.
J.Biochem., 167, 2020
|
|
5WR7
 
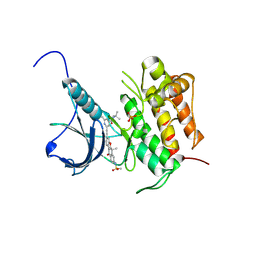 | | Crystal structure of Trk-A complexed with a selective inhibitor CH7057288 | | Descriptor: | High affinity nerve growth factor receptor, N-tert-butyl-2-[2-[6,6-dimethyl-8-(methylsulfonylamino)-11-oxidanylidene-naphtho[2,3-b][1]benzofuran-3-yl]ethynyl]-6-methyl-pyridine-4-carboxamide | | Authors: | Tanaka, H, Blaesse, M, Augustin, M, Goesser, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective TRK Inhibitor CH7057288 against TRK Fusion-Driven Cancer.
Mol. Cancer Ther., 17, 2018
|
|
9J6J
 
 | | HBx fused DDB1 4M mutant | | Descriptor: | Protein X,DNA damage-binding protein 1 | | Authors: | Tanaka, H, Kita, S, Sasaki, M, Maenaka, K, Machida, S. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the hepatitis B virus X protein in complex with DDB1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1IOI
 
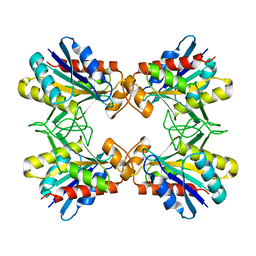 | | x-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, pyrococcus furiosus, and its cys-free mutant | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
1IOF
 
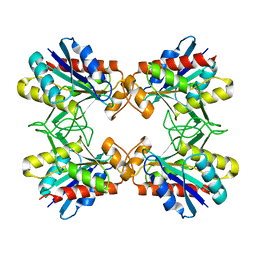 | | X-RAY CRYSTALLINE STRUCTURES OF PYRROLIDONE CARBOXYL PEPTIDASE FROM A HYPERTHERMOPHILE, PYROCOCCUS FURIOSUS, AND ITS CYS-FREE MUTANT | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
3ANU
 
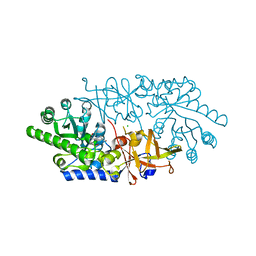 | | Crystal structure of D-serine dehydratase from chicken kidney | | Descriptor: | CHLORIDE ION, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
3VKF
 
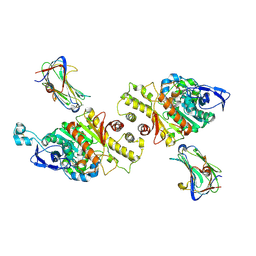 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
3ASI
 
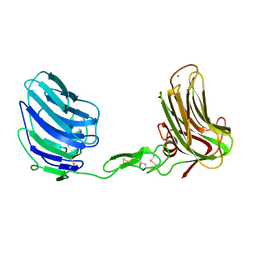 | | Alpha-Neurexin-1 ectodomain fragment; LNS5-EGF3-LNS6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-alpha | | Authors: | Tanaka, H, Nogi, T, Yasui, N, Takagi, J. | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Variant-Specific Neuroligin-Binding by alpha-Neurexin
Plos One, 6, 2011
|
|
3ANV
 
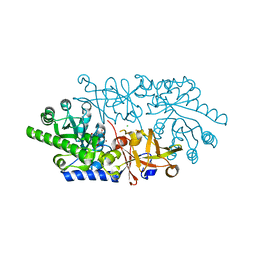 | | Crystal structure of D-serine dehydratase from chicken kidney (2,3-DAP complex) | | Descriptor: | 3-amino-D-alanine, CHLORIDE ION, D-serine dehydratase, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
3AWN
 
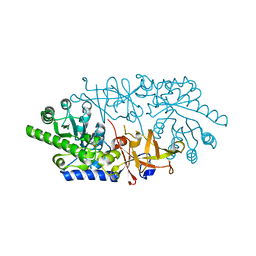 | | Crystal structure of D-serine dehydratase from chicken kidney (EDTA treated) | | Descriptor: | D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AWO
 
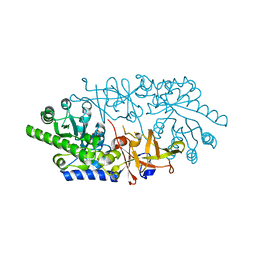 | | Crystal structure of D-serine dehydratase in complex with D-serine from chicken kidney (EDTA-treated) | | Descriptor: | D-SERINE, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
1AK6
 
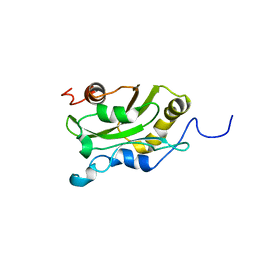 | | DESTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
1AK7
 
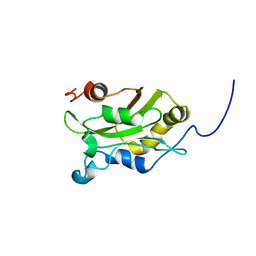 | | DESTRIN, NMR, 20 STRUCTURES | | Descriptor: | DESTRIN | | Authors: | Hatanaka, H, Moriyama, K, Ogura, K, Ichikawa, S, Yahara, I, Inagaki, F. | | Deposit date: | 1997-05-29 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of destrin and structural similarity between two actin-regulating protein families.
Cell(Cambridge,Mass.), 85, 1996
|
|
2CJN
 
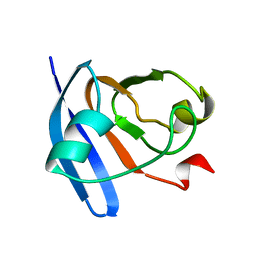 | | STRUCTURE OF FERREDOXIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
2CJO
 
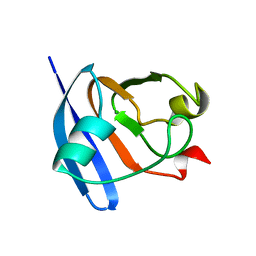 | | STRUCTURE OF FERREDOXIN, NMR, 10 STRUCTURES | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
1FRA
 
 | |
1ERA
 
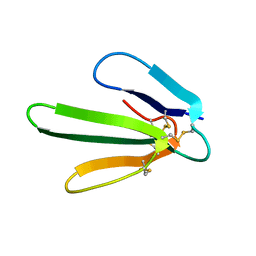 | |
1HEK
 
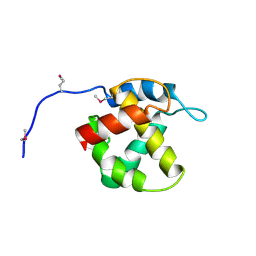 | | Crystal structure of equine infectious anaemia virus matrix antigen (EIAV MA) | | Descriptor: | GAG POLYPROTEIN, CORE PROTEIN P15 | | Authors: | Hatanaka, H, Iourin, O, Rao, Z, Fry, E, Kingsman, A, Stuart, D.I. | | Deposit date: | 2000-11-24 | | Release date: | 2001-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Equine Infectious Anemia Virus Matrix Protein.
J.Virol., 76, 2002
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
8Y3F
 
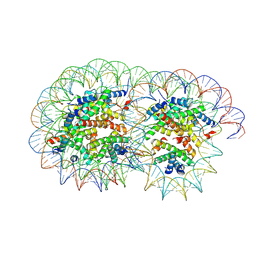 | | Cryo-EM structure of the overlapping di-nucleosome (intermediate form1) | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tanaka, H, Nishimura, M, Takizawa, Y, Nozawa, K, Ehara, H, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-01-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Asymmetric fluctuation of overlapping dinucleosome studied by cryoelectron microscopy and small-angle X-ray scattering.
Pnas Nexus, 3, 2024
|
|
8Y3D
 
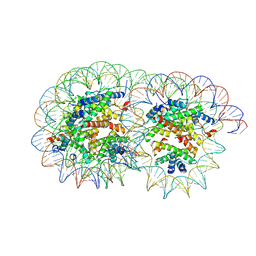 | | Cryo-EM structure of the overlapping di-nucleosome (intermediate form2) | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tanaka, H, Nishimura, M, Takizawa, Y, Nozawa, K, Ehara, H, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-01-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Asymmetric fluctuation of overlapping dinucleosome studied by cryoelectron microscopy and small-angle X-ray scattering.
Pnas Nexus, 3, 2024
|
|
8Y3C
 
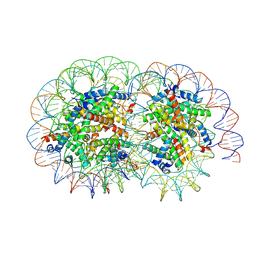 | | Cryo-EM structure of the overlapping di-nucleosome (closed form) | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tanaka, H, Nishimura, M, Takizawa, Y, Nozawa, K, Ehara, H, Sekine, S, Kurumizaka, H. | | Deposit date: | 2024-01-29 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (5.21 Å) | | Cite: | Asymmetric fluctuation of overlapping dinucleosome studied by cryoelectron microscopy and small-angle X-ray scattering.
Pnas Nexus, 3, 2024
|
|
