2SOD
 
 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
1UGH
 
 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
4TWS
 
 | | Gadolinium Derivative of Tetragonal Hen Egg-White Lysozyme at 1.45 A Resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Holton, J.M, Classen, S, Frankel, K.A, Tainer, J.A. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The R-factor gap in macromolecular crystallography: an untapped potential for insights on accurate structures.
Febs J., 281, 2014
|
|
1UGI
 
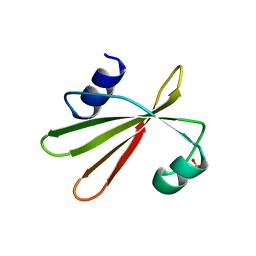 | | URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | IMIDAZOLE, SULFATE ION, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
3SOK
 
 | | Dichelobacter nodosus pilin FimA | | Descriptor: | Fimbrial protein | | Authors: | Arvai, A.S, Craig, L, Hartung, S, Wood, T, Kolappan, S, Shin, D.S, Tainer, J.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
7N8W
 
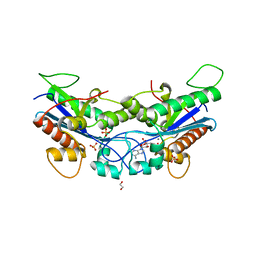 | |
7N8V
 
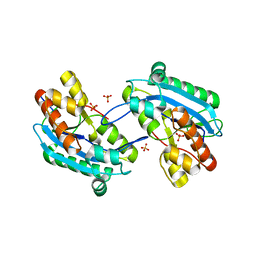 | |
3CRW
 
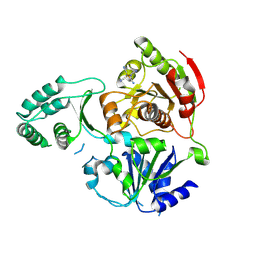 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3CRV
 
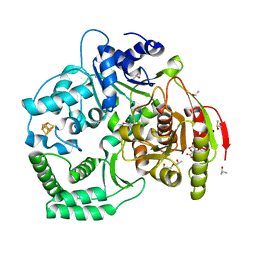 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
8TUK
 
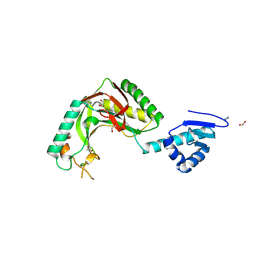 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | Descriptor: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | Authors: | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | Deposit date: | 2023-08-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
8TLY
 
 | |
4YEX
 
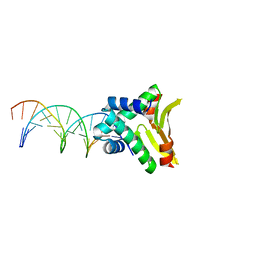 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFT
 
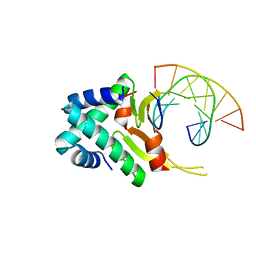 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEW
 
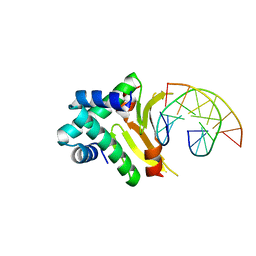 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEY
 
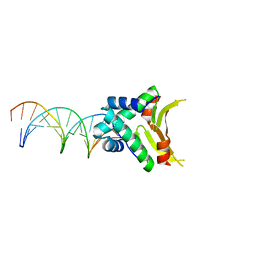 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
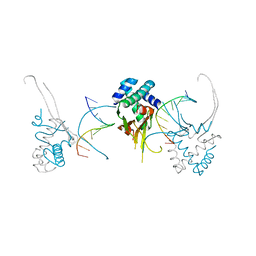 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YDD
 
 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
4YDS
 
 | |
5UM9
 
 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
1KYR
 
 | | Crystal Structure of a Cu-bound Green Fluorescent Protein Zn Biosensor | | Descriptor: | COPPER (II) ION, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
8VGY
 
 | | Crystal structure of human apoptosis-inducing factor (AIF) bound to the fused N-terminal domain of CHCHD4 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis-inducing factor 1, mitochondrial,Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Brosey, C.A, Tainer, J.A. | | Deposit date: | 2023-12-29 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
Embo J., 44, 2025
|
|
8GJ9
 
 | | RAD51C N-terminal domain | | Descriptor: | RAD51C, ZINC ION | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8GJA
 
 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
