5AXH
 
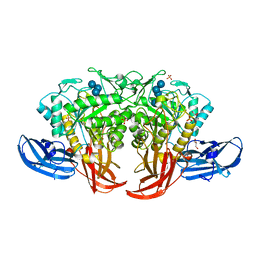 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
5AXG
 
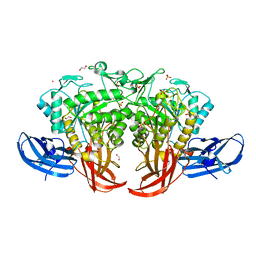 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Dextranase, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
3WNO
 
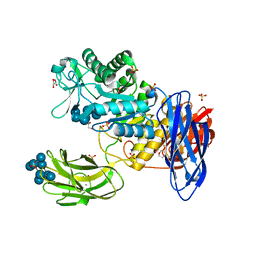 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNM
 
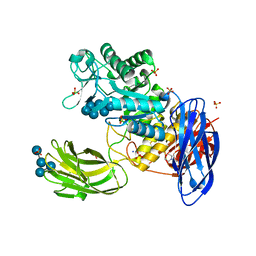 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNL
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNK
 
 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNN
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3AI5
 
 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1X2W
 
 | | Crystal Structure of Apo-Habu IX-bp at pH 4.6 | | Descriptor: | CHLORIDE ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
1X2T
 
 | | Crystal Structure of Habu IX-bp at pH 6.5 | | Descriptor: | CALCIUM ION, Coagulation factor IX/X-binding protein A chain, Coagulation factor IX/factor X-binding protein B chain, ... | | Authors: | Suzuki, N, Fujimoto, Z, Morita, T, Fukamizu, A, Mizuno, H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | pH-Dependent Structural Changes at Ca(2+)-binding sites of Coagulation Factor IX-binding Protein
J.Mol.Biol., 353, 2005
|
|
3VMP
 
 | | Crystal structure of dextranase from Streptococcus mutans in complex with 4,5-epoxypentyl alpha-D-glucopyranoside | | Descriptor: | 5-hydroxypentyl alpha-D-glucopyranoside, Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
3VMN
 
 | | Crystal structure of dextranase from Streptococcus mutans | | Descriptor: | Dextranase, PHOSPHATE ION | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
3VMO
 
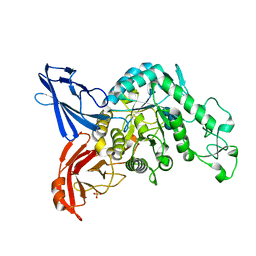 | | Crystal structure of dextranase from Streptococcus mutans in complex with isomaltotriose | | Descriptor: | Dextranase, PHOSPHATE ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Okuyama, M, Mori, H, Funane, K, Kimura, A. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of dextran degradation mechanism by streptococcus mutans dextranase belonging to glycoside hydrolase family 66
J.Biol.Chem., 287, 2012
|
|
2DDB
 
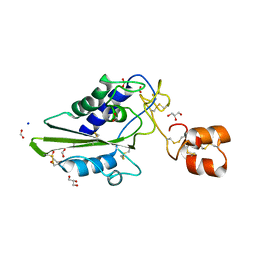 | | Crystal structure of pseudecin from Pseudechis porphyriacus | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudecin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2DDA
 
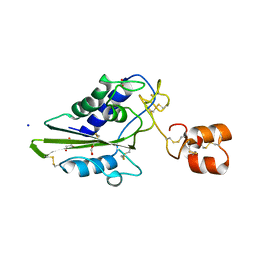 | | Crystal structure of pseudechetoxin from Pseudechis australis | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudechetoxin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EPF
 
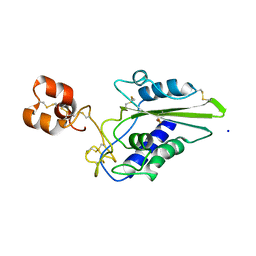 | | Crystal Structure of Zinc-Bound Pseudecin From Pseudechis Porphyriacus | | Descriptor: | Pseudecin, SODIUM ION, ZINC ION | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2007-03-29 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VHT
 
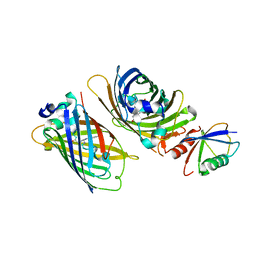 | | Crystal structure of GFP-Wrnip1 UBZ domain fusion protein in complex with ubiquitin | | Descriptor: | Green fluorescent protein, Green fluorescent protein,ATPase WRNIP1, Ubiquitin, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2011-09-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 2016
|
|
3VHS
 
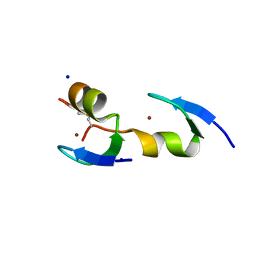 | |
3WUP
 
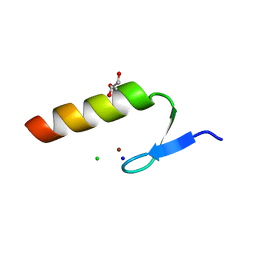 | | Crystal Structure of the Ubiquitin-Binding Zinc Finger (UBZ) Domain of the Human DNA Polymerase Eta | | Descriptor: | CHLORIDE ION, DNA polymerase eta, GLYCEROL, ... | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, S. | | Deposit date: | 2014-05-01 | | Release date: | 2015-06-17 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Febs J., 283, 2016
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6H
 
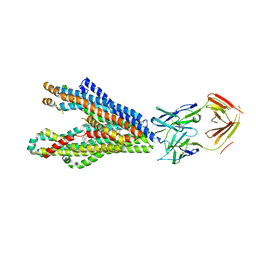 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
5X7G
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
5ND1
 
 | | Viral evolution results in multiple, surface-allocated enzymatic activities in a fungal double-stranded RNA virus | | Descriptor: | Capsid protein | | Authors: | Mata, C.P, Luque, D, Gomez Blanco, J, Rodriguez, J.M, Suzuki, N, Ghabrial, S.A, Carrascosa, J.L, Trus, B.L, Caston, J.R. | | Deposit date: | 2017-03-07 | | Release date: | 2017-11-29 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Acquisition of functions on the outer capsid surface during evolution of double-stranded RNA fungal viruses.
PLoS Pathog., 13, 2017
|
|
5X7S
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
