1GYT
 
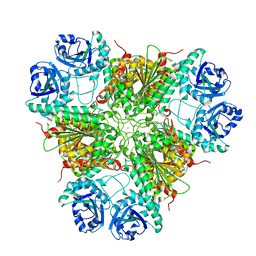 | | E. coli Aminopeptidase A (PepA) | | Descriptor: | CARBONATE ION, CHLORIDE ION, CYTOSOL AMINOPEPTIDASE, ... | | Authors: | Straeter, N. | | Deposit date: | 2002-04-29 | | Release date: | 2002-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of Aminopeptidase a from Escherichia Coli and a Model for the Nucleoprotein Complex in Xer Site-Specific Recombination
Embo J., 18, 1999
|
|
1LAN
 
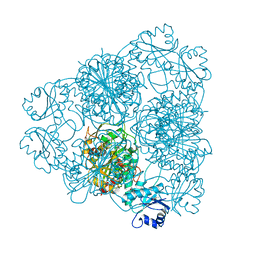 | | LEUCINE AMINOPEPTIDASE COMPLEX WITH L-LEUCINAL | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEUCINE, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LAM
 
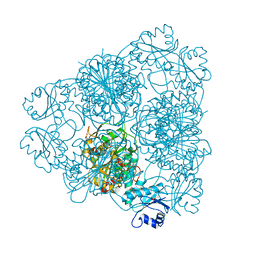 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1HO5
 
 | |
1OID
 
 | |
1OI8
 
 | | 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, PROTEIN USHA, ... | | Authors: | Schultz-Heienbrok, R, Maier, T, Straeter, N. | | Deposit date: | 2003-06-10 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trapping a 96 Degree Domain Rotation in Two Distinct Conformations by Engineered Disulfide Bridges
Protein Sci., 13, 2004
|
|
1OIE
 
 | |
1HP1
 
 | | 5'-NUCLEOTIDASE (OPEN FORM) COMPLEX WITH ATP | | Descriptor: | 5'-NUCLEOTIDASE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-12 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
1HPU
 
 | | 5'-NUCLEOTIDASE (CLOSED FORM), COMPLEX WITH AMPCP | | Descriptor: | 5'-NUCLEOTIDASE, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-13 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
4U1R
 
 | |
4K7L
 
 | | Crystal structure of RNase S variant (K7C/Q11C) | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of RNase S with a [Hg(Cys2)] metal center in the S-peptide as a template for structure-based design of artificial metalloenzymes using peptide-protein complementation
To be Published
|
|
4K7M
 
 | | Crystal structure of RNase S variant (K7C/Q11C) with bound mercury ions | | Descriptor: | MERCURY (II) ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo- and Metalated Thiolate containing RNase S as Structural Basis for the Design of Artificial Metalloenzymes by Peptide- Protein Complementation
Z.Anorg.Allg.Chem., 639, 2013
|
|
4EZU
 
 | |
4EZS
 
 | |
4EZV
 
 | |
3ZNU
 
 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZNJ
 
 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZO7
 
 | | Crystal structure of ClcFE27A with substrate | | Descriptor: | (2S)-2-chloranyl-2-[(2R)-5-oxidanylidene-2H-furan-2-yl]ethanoic acid, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
4A57
 
 | |
4A59
 
 | |
4A5A
 
 | |
4BR5
 
 | | Rat NTPDase2 in complex with Zn AMPPNP | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRO
 
 | |
4BQZ
 
 | | Rat NTPDase2 in complex with Mg GMPPNP | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRD
 
 | | Legionella pneumophila NTPDase1 Q193E crystal form II, closed, Mg AMPPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
